Bicc1 Polymerization Regulates the Localization and Silencing of Bound mRNA
- PMID: 26217012
- PMCID: PMC4561730
- DOI: 10.1128/MCB.00341-15
Bicc1 Polymerization Regulates the Localization and Silencing of Bound mRNA
Abstract
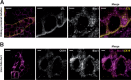
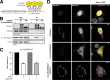
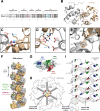
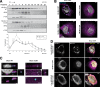
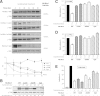
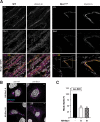
Loss of the RNA-binding protein Bicaudal-C (Bicc1) provokes renal and pancreatic cysts as well as ectopic Wnt/β-catenin signaling during visceral left-right patterning. Renal cysts are linked to defective silencing of Bicc1 target mRNAs, including adenylate cyclase 6 (AC6). RNA binding of Bicc1 is mediated by N-terminal KH domains, whereas a C-terminal sterile alpha motif (SAM) self-polymerizes in vitro and localizes Bicc1 in cytoplasmic foci in vivo. To assess a role for multimerization in silencing, we conducted structure modeling and then mutated the SAM domain residues which in this model were predicted to polymerize Bicc1 in a left-handed helix. We show that a SAM-SAM interface concentrates Bicc1 in cytoplasmic clusters to specifically localize and silence bound mRNA. In addition, defective polymerization decreases Bicc1 stability and thus indirectly attenuates inhibition of Dishevelled 2 in the Wnt/β-catenin pathway. Importantly, aberrant C-terminal extension of the SAM domain in bpk mutant Bicc1 phenocopied these defects. We conclude that polymerization is a novel disease-relevant mechanism both to stabilize Bicc1 and to present associated mRNAs in specific silencing platforms.
Copyright © 2015, Rothé et al.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
