Transcription Factor T-Bet in Atlantic Salmon: Characterization and Gene Expression in Mucosal Tissues during Aeromonas Salmonicida Infection
- PMID: 26217339
- PMCID: PMC4492157
- DOI: 10.3389/fimmu.2015.00345
Transcription Factor T-Bet in Atlantic Salmon: Characterization and Gene Expression in Mucosal Tissues during Aeromonas Salmonicida Infection
Abstract
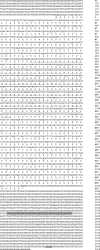
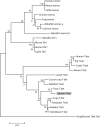
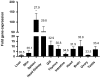
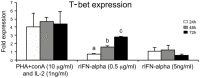
The T-box transcription factor T-bet is expressed in a number of hematopoietic cell types in mammals and plays an essential role in the lineage determination of Th1 T-helper cells and is considered as an essential feature for both innate and adaptive immune responses in higher vertebrates. In the present study, we have identified and characterized the full-length Atlantic salmon T-bet cDNA (3502 bp). The putative primary structure of the polypeptide deduced from the cDNA sequence contained 612 aa, which possessed a T-box DNA binding domain. Phylogenetic study and gene synteny revealed it is as a homolog to mammalian T-bet. Quantitative PCR analysis of different tissues in healthy fish showed that salmon T-bet gene was highly expressed in spleen, followed by head kidney, and was expressed in intestine, skin, and liver at lower levels. Moreover, the time-dependent expression profile of T-bet, interferon gamma (IFNγ), interleukin-22 (IL-22), and natural killer enhancement factor in mucosal tissues during water-borne infection with live Aeromonas salmonicida, indicated the involvement of T-bet in mucosal immune response in Atlantic salmon.
Keywords: A. salmonicida; Atlantic salmon; T-bet; gene expression; mucosal immunity.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

