Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula
- PMID: 26217660
- PMCID: PMC4500099
- DOI: 10.3389/fbioe.2015.00097
Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula
Abstract
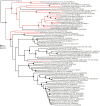
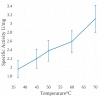
Mercuric ion reductase (MerA), a mercury detoxification enzyme, has been tuned by evolution to have high specificity for mercuric ions (Hg(2+)) and to catalyze their reduction to a more volatile, less toxic elemental form. Here, we present a biochemical and structural characterization of MerA from the thermophilic crenarchaeon Metallosphaera sedula. MerA from M. sedula is a thermostable enzyme, and remains active after extended incubation at 97°C. At 37°C, the NADPH oxidation-linked Hg(2+) reduction specific activity was found to be 1.9 μmol/min⋅mg, increasing to 3.1 μmol/min⋅mg at 70°C. M. sedula MerA crystals were obtained and the structure was solved to 1.6 Å, representing the first solved crystal structure of a thermophilic MerA. Comparison of both the crystal structure and amino acid sequence of MerA from M. sedula to mesophillic counterparts provides new insights into the structural determinants that underpin the thermal stability of the enzyme.
Keywords: MerA; biosensor; mercuric reductase; mercury detoxification; structure; thermophile; thermostability.
Figures
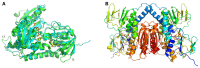
Similar articles
-
Thermal Stability of a Mercuric Reductase from the Red Sea Atlantis II Hot Brine Environment as Analyzed by Site-Directed Mutagenesis.Appl Environ Microbiol. 2019 Jan 23;85(3):e02387-18. doi: 10.1128/AEM.02387-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30446558 Free PMC article.
-
Mercury resistance and mercuric reductase activities and expression among chemotrophic thermophilic Aquificae.Appl Environ Microbiol. 2012 Sep;78(18):6568-75. doi: 10.1128/AEM.01060-12. Epub 2012 Jul 6. Appl Environ Microbiol. 2012. PMID: 22773655 Free PMC article.
-
A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase.Environ Microbiol. 2010 Nov;12(11):2904-17. doi: 10.1111/j.1462-2920.2010.02260.x. Environ Microbiol. 2010. PMID: 20545753
-
Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts.FASEB J. 1988 Feb;2(2):124-30. doi: 10.1096/fasebj.2.2.3277886. FASEB J. 1988. PMID: 3277886 Review.
-
Microbes in mercury-enriched geothermal springs in western North America.Sci Total Environ. 2016 Nov 1;569-570:321-331. doi: 10.1016/j.scitotenv.2016.06.080. Epub 2016 Jun 24. Sci Total Environ. 2016. PMID: 27344121 Review.
Cited by
-
Temporal dynamics in a red alga dominated geothermal feature in Yellowstone National Park.ISME Commun. 2024 Dec 3;4(1):ycae151. doi: 10.1093/ismeco/ycae151. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39711979 Free PMC article.
-
Enzyme Engineering: Performance Optimization, Novel Sources, and Applications in the Food Industry.Foods. 2024 Nov 28;13(23):3846. doi: 10.3390/foods13233846. Foods. 2024. PMID: 39682920 Free PMC article. Review.
-
Thermal Stability of a Mercuric Reductase from the Red Sea Atlantis II Hot Brine Environment as Analyzed by Site-Directed Mutagenesis.Appl Environ Microbiol. 2019 Jan 23;85(3):e02387-18. doi: 10.1128/AEM.02387-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30446558 Free PMC article.
-
Expanded Diversity and Phylogeny of mer Genes Broadens Mercury Resistance Paradigms and Reveals an Origin for MerA Among Thermophilic Archaea.Front Microbiol. 2021 Jun 23;12:682605. doi: 10.3389/fmicb.2021.682605. eCollection 2021. Front Microbiol. 2021. PMID: 34248899 Free PMC article.
-
Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.Sci Rep. 2023 Oct 19;13(1):17854. doi: 10.1038/s41598-023-44968-w. Sci Rep. 2023. PMID: 37857791 Free PMC article.
References
-
- Adami M., Martini M., Piras L. (1995). Characterization and enzymatic application of a redox potential biosensor based on a silicon transducer. Biosens. Bioelectron. 10, 633–638.10.1016/0956-5663(95)96939-V - DOI
-
- Auernik K. S., Maezato Y., Blum P. H., Kelly R. M. (2008b). The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74, 682–692.10.1128/aem.02019-07 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

