Mechanisms of hypervirulent Clostridium difficile ribotype 027 displacement of endemic strains: an epidemiological model
- PMID: 26218654
- PMCID: PMC4517512
- DOI: 10.1038/srep12666
Mechanisms of hypervirulent Clostridium difficile ribotype 027 displacement of endemic strains: an epidemiological model
Abstract
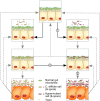
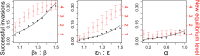
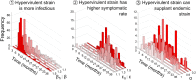
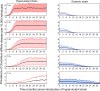
Following rapid, global clonal dominance of hypervirulent ribotypes, Clostridium difficile now constitutes the primary infectious cause of nosocomial diarrhoea. Evidence indicates at least three possible mechanisms of hypervirulence that facilitates the successful invasion of these atypical strains: 1) increased infectiousness relative to endemic strains; 2) increased symptomatic disease rate relative to endemic strains; and 3) an ability to outcompete endemic strains in the host's gut. Stochastic simulations of an infection transmission model demonstrate clear differences between the invasion potentials of C. difficile strains utilising the alternative hypervirulence mechanisms, and provide new evidence that favours certain mechanisms (1 and 2) more than others (3). Additionally, simulations illustrate that direct competition between strains (inside the host's gut) is not a prerequisite for the sudden switching that has been observed in prevailing ribotypes; previously dominant C. difficile strains can be excluded by hypervirulent ribotypes through indirect (exploitative) competition.
Figures
Similar articles
-
Sporulation properties and antimicrobial susceptibility in endemic and rare Clostridium difficile PCR ribotypes.Anaerobe. 2016 Jun;39:183-8. doi: 10.1016/j.anaerobe.2016.04.010. Epub 2016 Apr 14. Anaerobe. 2016. PMID: 27095618
-
Genetic markers for Clostridium difficile lineages linked to hypervirulence.Microbiology (Reading). 2011 Nov;157(Pt 11):3113-3123. doi: 10.1099/mic.0.051953-0. Epub 2011 Aug 26. Microbiology (Reading). 2011. PMID: 21873406
-
Molecular epidemiology of endemic Clostridium difficile infection and the significance of subtypes of the United Kingdom epidemic strain (PCR ribotype 1).J Clin Microbiol. 2005 Jun;43(6):2685-96. doi: 10.1128/JCM.43.6.2685-2696.2005. J Clin Microbiol. 2005. PMID: 15956384 Free PMC article.
-
New trends in Clostridium difficile virulence and pathogenesis.Int J Antimicrob Agents. 2009 Mar;33 Suppl 1:S24-8. doi: 10.1016/S0924-8579(09)70012-3. Int J Antimicrob Agents. 2009. PMID: 19303565 Review.
-
Ribotypes and New Virulent Strains Across Europe.Adv Exp Med Biol. 2018;1050:45-58. doi: 10.1007/978-3-319-72799-8_4. Adv Exp Med Biol. 2018. PMID: 29383663 Review.
Cited by
-
Effect of Bifidobacterium upon Clostridium difficile Growth and Toxicity When Co-cultured in Different Prebiotic Substrates.Front Microbiol. 2016 May 18;7:738. doi: 10.3389/fmicb.2016.00738. eCollection 2016. Front Microbiol. 2016. PMID: 27242753 Free PMC article.
-
An in silico evaluation of treatment regimens for recurrent Clostridium difficile infection.PLoS One. 2017 Aug 11;12(8):e0182815. doi: 10.1371/journal.pone.0182815. eCollection 2017. PLoS One. 2017. PMID: 28800598 Free PMC article.
-
A global to local genomics analysis of Clostridioides difficile ST1/RT027 identifies cryptic transmission events in a northern Arizona healthcare network.Microb Genom. 2019 Jul;5(7):e000271. doi: 10.1099/mgen.0.000271. Epub 2019 May 20. Microb Genom. 2019. PMID: 31107202 Free PMC article.
-
Aryl-alkyl-lysines: Novel agents for treatment of C. difficile infection.Sci Rep. 2020 Mar 27;10(1):5624. doi: 10.1038/s41598-020-62496-9. Sci Rep. 2020. PMID: 32221399 Free PMC article.
-
Mathematical modelling for antibiotic resistance control policy: do we know enough?BMC Infect Dis. 2019 Nov 29;19(1):1011. doi: 10.1186/s12879-019-4630-y. BMC Infect Dis. 2019. PMID: 31783803 Free PMC article.
References
-
- Kelly C. P. & LaMont J. T. Clostridium difficile—more difficult than ever. New Engl. J. Med. 359, 1932–1940 (2008). - PubMed
-
- Barbut F. et al. Prospective study of Clostridium difficile-associated disease in Europe with phenotypic and genotypic characterization of the isolates. Clin. Microbiol. Infect. 13, 1048–1057 (2007). - PubMed
-
- Kuijper E. J. et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe. Eurosurveil. 13, e18942 (2008). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

