Learning the Structure of Biomedical Relationships from Unstructured Text
- PMID: 26219079
- PMCID: PMC4517797
- DOI: 10.1371/journal.pcbi.1004216
Learning the Structure of Biomedical Relationships from Unstructured Text
Abstract
The published biomedical research literature encompasses most of our understanding of how drugs interact with gene products to produce physiological responses (phenotypes). Unfortunately, this information is distributed throughout the unstructured text of over 23 million articles. The creation of structured resources that catalog the relationships between drugs and genes would accelerate the translation of basic molecular knowledge into discoveries of genomic biomarkers for drug response and prediction of unexpected drug-drug interactions. Extracting these relationships from natural language sentences on such a large scale, however, requires text mining algorithms that can recognize when different-looking statements are expressing similar ideas. Here we describe a novel algorithm, Ensemble Biclustering for Classification (EBC), that learns the structure of biomedical relationships automatically from text, overcoming differences in word choice and sentence structure. We validate EBC's performance against manually-curated sets of (1) pharmacogenomic relationships from PharmGKB and (2) drug-target relationships from DrugBank, and use it to discover new drug-gene relationships for both knowledge bases. We then apply EBC to map the complete universe of drug-gene relationships based on their descriptions in Medline, revealing unexpected structure that challenges current notions about how these relationships are expressed in text. For instance, we learn that newer experimental findings are described in consistently different ways than established knowledge, and that seemingly pure classes of relationships can exhibit interesting chimeric structure. The EBC algorithm is flexible and adaptable to a wide range of problems in biomedical text mining.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
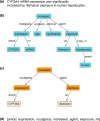
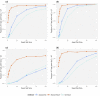


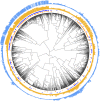
References
-
- http://www.nlm.nih.gov/bsd/num_titles.html. Accessed 3/3/14.
-
- http://www.nlm.nih.gov/bsd/medline_cit_counts_yr_pub.html. Accessed 3/3/14.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

