Extracellular milieu grossly alters pathogen-specific immune response of mammary epithelial cells
- PMID: 26219462
- PMCID: PMC4518681
- DOI: 10.1186/s12917-015-0489-3
Extracellular milieu grossly alters pathogen-specific immune response of mammary epithelial cells
Abstract
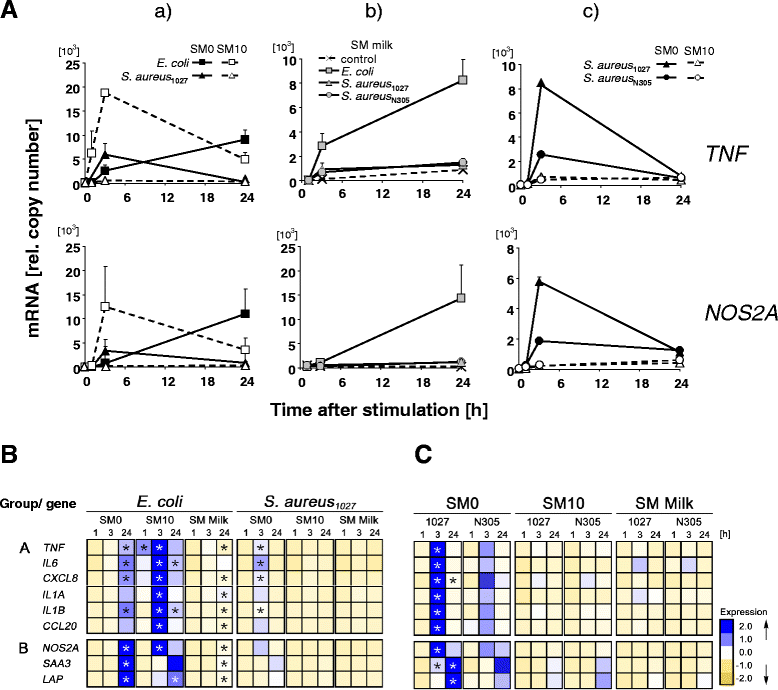
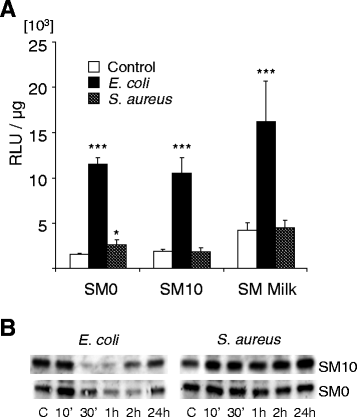
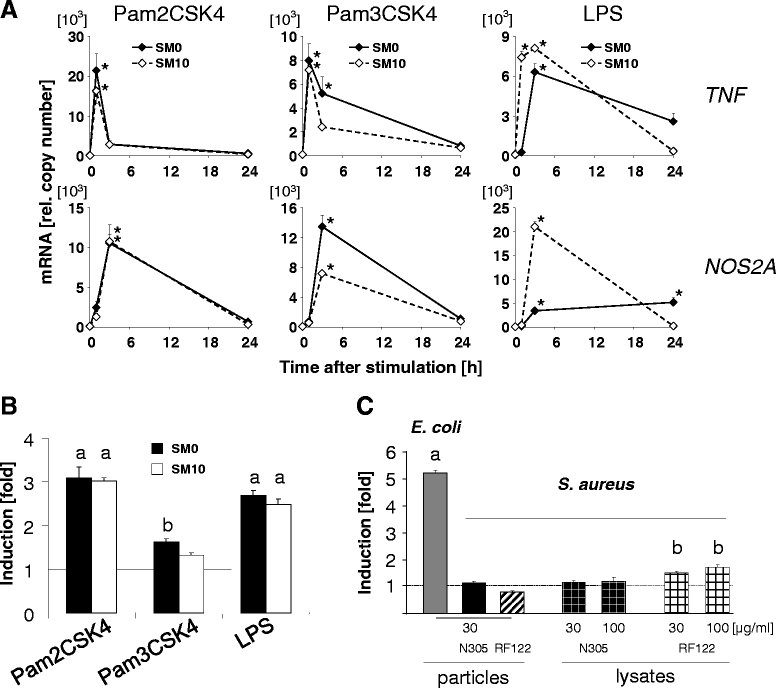
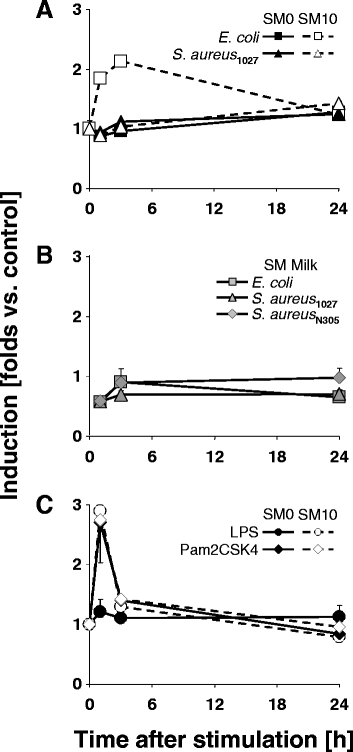
Background: Considerably divergent data have been published from attempts to model the E. coli vs. S. aureus specific immune reaction of the udder using primary cultures of bovine mammary epithelial cells from cows (pbMEC). Some groups reported a swift, strong and transient inflammatory response against challenges with E. coli and only a weak and retarded response against S. aureus, in agreement with the respective reaction of the udder. Others found almost the reverse. Presence or absence of fetal calf serum distinguished the experimental setting between both groups. We examined here if this causes the divergent reaction of the pbMEC towards both pathogen species. We challenged pbMEC with proteins from heat killed E. coli or S. aureus pathogens or purified TLR2 and TLR4 ligands. The stimuli were applied in normal growth medium with (SM10) or without (SM0) 10% fetal calf serum, or in the basal medium supplemented with 10 mg/ml milk proteins (SM Milk).
Results: Withdrawal of FCS slowed down and decreased the extent by which E. coli or LPS enhanced the expression of cyto- and chemokine encoding genes through impaired TLR4 signalling but enforced their expression during stimulation with S. aureus. SM Milk strongly quenched the induction of those genes. S. aureus strain specific differences in the reaction of the pbMEC could only be recorded in SM0. NF-κB factors were activated by E. coli in all stimulation media, but only to a small extent by S. aureus, solely in SM0. Purified ligands for TLR2 stimulated expression of those genes and activated NF-κB equally well in SM10 and SM0. The mRNA destabilizing factor tristetraproline was only induced by E. coli in SM10 and by purified PAMPs.
Conclusions: Our data cross validate the correctness of previously published divergent data on the pathogen-specific induction of key immune genes in pbMEC. The differences are due to the presence of FCS, modulating signalling through TLR4 and TLR-unrelated pathogen receptors. S. aureus does not substantially activate any TLR signalling in MEC. Rather, receptors distinct from TLRs perceive the presence of S. aureus and control the immune response against this pathogen in MEC.
Figures
Similar articles
-
Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-kappaB in mammary epithelial cells and to quickly induce TNFalpha and interleukin-8 (CXCL8) expression in the udder.Mol Immunol. 2008 Mar;45(5):1385-97. doi: 10.1016/j.molimm.2007.09.004. Epub 2007 Oct 22. Mol Immunol. 2008. PMID: 17936907
-
Staphylococcus aureus and Escherichia coli elicit different innate immune responses from bovine mammary epithelial cells.Vet Immunol Immunopathol. 2013 Oct 1;155(4):245-52. doi: 10.1016/j.vetimm.2013.08.003. Epub 2013 Aug 24. Vet Immunol Immunopathol. 2013. PMID: 24018311
-
NF-kappaB factors are essential, but not the switch, for pathogen-related induction of the bovine beta-defensin 5-encoding gene in mammary epithelial cells.Mol Immunol. 2006 Feb;43(3):210-25. doi: 10.1016/j.molimm.2005.02.003. Mol Immunol. 2006. PMID: 16199258
-
Bacterial lipopolysaccharide induces increased expression of toll-like receptor (TLR) 4 and downstream TLR signaling molecules in bovine mammary epithelial cells.Vet Res. 2008 Mar-Apr;39(2):11. doi: 10.1051/vetres:2007047. Epub 2007 Dec 21. Vet Res. 2008. PMID: 18096120
-
Expression, Regulation, and Function of β-Defensins in the Bovine Mammary Glands: Current Knowledge and Future Perspectives.Animals (Basel). 2023 Oct 31;13(21):3372. doi: 10.3390/ani13213372. Animals (Basel). 2023. PMID: 37958127 Free PMC article. Review.
Cited by
-
Differentiating Staphylococcus aureus from Escherichia coli mastitis: S. aureus triggers unbalanced immune-dampening and host cell invasion immediately after udder infection.Sci Rep. 2017 Jul 6;7(1):4811. doi: 10.1038/s41598-017-05107-4. Sci Rep. 2017. PMID: 28684793 Free PMC article.
-
Comparison of the pathogen species-specific immune response in udder derived cell types and their models.Vet Res. 2016 Feb 1;47:22. doi: 10.1186/s13567-016-0307-3. Vet Res. 2016. PMID: 26830914 Free PMC article.
-
Immune defenses of the mammary gland epithelium of dairy ruminants.Front Immunol. 2022 Oct 21;13:1031785. doi: 10.3389/fimmu.2022.1031785. eCollection 2022. Front Immunol. 2022. PMID: 36341445 Free PMC article. Review.
-
Escherichia coli and Staphylococcus aureus Differentially Regulate Nrf2 Pathway in Bovine Mammary Epithelial Cells: Relation to Distinct Innate Immune Response.Cells. 2021 Dec 6;10(12):3426. doi: 10.3390/cells10123426. Cells. 2021. PMID: 34943933 Free PMC article.
-
The in vitro host cell immune response to bovine-adapted Staphylococcus aureus varies according to bacterial lineage.Sci Rep. 2019 Apr 16;9(1):6134. doi: 10.1038/s41598-019-42424-2. Sci Rep. 2019. PMID: 30992458 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

