A roadmap for the genetic analysis of renal aging
- PMID: 26219736
- PMCID: PMC4568960
- DOI: 10.1111/acel.12378
A roadmap for the genetic analysis of renal aging
Abstract
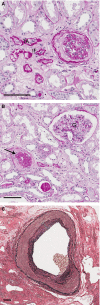
Several studies show evidence for the genetic basis of renal disease, which renders some individuals more prone than others to accelerated renal aging. Studying the genetics of renal aging can help us to identify genes involved in this process and to unravel the underlying pathways. First, this opinion article will give an overview of the phenotypes that can be observed in age-related kidney disease. Accurate phenotyping is essential in performing genetic analysis. For kidney aging, this could include both functional and structural changes. Subsequently, this article reviews the studies that report on candidate genes associated with renal aging in humans and mice. Several loci or candidate genes have been found associated with kidney disease, but identification of the specific genetic variants involved has proven to be difficult. CUBN, UMOD, and SHROOM3 were identified by human GWAS as being associated with albuminuria, kidney function, and chronic kidney disease (CKD). These are promising examples of genes that could be involved in renal aging, and were further mechanistically evaluated in animal models. Eventually, we will provide approaches for performing genetic analysis. We should leverage the power of mouse models, as testing in humans is limited. Mouse and other animal models can be used to explain the underlying biological mechanisms of genes and loci identified by human GWAS. Furthermore, mouse models can be used to identify genetic variants associated with age-associated histological changes, of which Far2, Wisp2, and Esrrg are examples. A new outbred mouse population with high genetic diversity will facilitate the identification of genes associated with renal aging by enabling high-resolution genetic mapping while also allowing the control of environmental factors, and by enabling access to renal tissues at specific time points for histology, proteomics, and gene expression.
Keywords: aging; genetics; human; kidney; mouse; phenotype.
© 2015 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures
References
-
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. - PubMed
-
- Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, Yu R, Jonker JW, Holt JA, Rajpal DK, Li H, Stuart J, McPherson R, Remlinger KS, Chang CY, McDonnell DP, Evans RM, Billin AN. ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 2010;24:299–309. - PMC - PubMed
-
- Anderson S, Brenner BM. Effects of aging on the renal glomerulus. Am. J. Med. 1986;80:435–442. - PubMed
-
- Boger CA, Chen MH, Tin A, Olden M, Kottgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tonjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarod K, Skorpen F, Syvanen AC, Illig T, Baumert J, Koenig W, Kramer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Volzke H, Stumvoll M, Magi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlov J, Hallan S, Navis G, CKDGen Consortium, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH. CUBN is a gene locus for albuminuria. J. Am. Soc. Nephrol. 2011;22:555–570. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

