CD161(int)CD8+ T cells: a novel population of highly functional, memory CD8+ T cells enriched within the gut
- PMID: 26220166
- PMCID: PMC4732939
- DOI: 10.1038/mi.2015.69
CD161(int)CD8+ T cells: a novel population of highly functional, memory CD8+ T cells enriched within the gut
Abstract
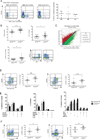
The C-type lectin-like receptor CD161 is expressed by lymphocytes found in human gut and liver, as well as blood, especially natural killer (NK) cells, T helper 17 (Th17) cells, and a population of unconventional T cells known as mucosal-associated invariant T (MAIT) cells. The association of high CD161 expression with innate T-cell populations including MAIT cells is established. Here we show that CD161 is also expressed, at intermediate levels, on a prominent subset of polyclonal CD8+ T cells, including antiviral populations that display a memory phenotype. These memory CD161(int)CD8+ T cells are enriched within the colon and express both CD103 and CD69, markers associated with tissue residence. Furthermore, this population was characterized by enhanced polyfunctionality, increased levels of cytotoxic mediators, and high expression of the transcription factors T-bet and eomesodermin (EOMES). Such populations were induced by novel vaccine strategies based on adenoviral vectors, currently in trial against hepatitis C virus. Thus, intermediate CD161 expression marks potent polyclonal, polyfunctional tissue-homing CD8+ T-cell populations in humans. As induction of such responses represents a major aim of T-cell prophylactic and therapeutic vaccines in viral disease and cancer, analysis of these populations could be of value in the future.
Conflict of interest statement
Disclosure: The authors declare no conflict of interest.
Figures




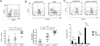
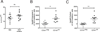
References
-
- Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 2009;10:524–530. - PubMed
-
- Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting Edge: Gut Microenvironment Promotes Differentiation of a Unique Memory CD8 T Cell Population. J. Immunol. 2006;176:2079–2083. - PubMed
-
- Lanier LL, Chang C, Phillips JH. Human NKRP1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J. Immunol. 1994;153:2417–2428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

