Network analysis of temporal functionalities of the gut induced by perturbations in new-born piglets
- PMID: 26220188
- PMCID: PMC4518884
- DOI: 10.1186/s12864-015-1733-8
Network analysis of temporal functionalities of the gut induced by perturbations in new-born piglets
Abstract
Background: Evidence is accumulating that perturbation of early life microbial colonization of the gut induces long-lasting adverse health effects in individuals. Understanding the mechanisms behind these effects will facilitate modulation of intestinal health. The objective of this study was to identify biological processes involved in these long lasting effects and the (molecular) factors that regulate them. We used an antibiotic and the same antibiotic in combination with stress on piglets as an early life perturbation. Then we used host gene expression data from the gut (jejunum) tissue and community-scale analysis of gut microbiota from the same location of the gut, at three different time-points to gauge the reaction to the perturbation. We analysed the data by a new combination of existing tools. First, we analysed the data in two dimensions, treatment and time, with quadratic regression analysis. Then we applied network-based data integration approaches to find correlations between host gene expression and the resident microbial species.
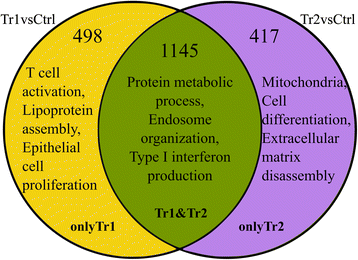
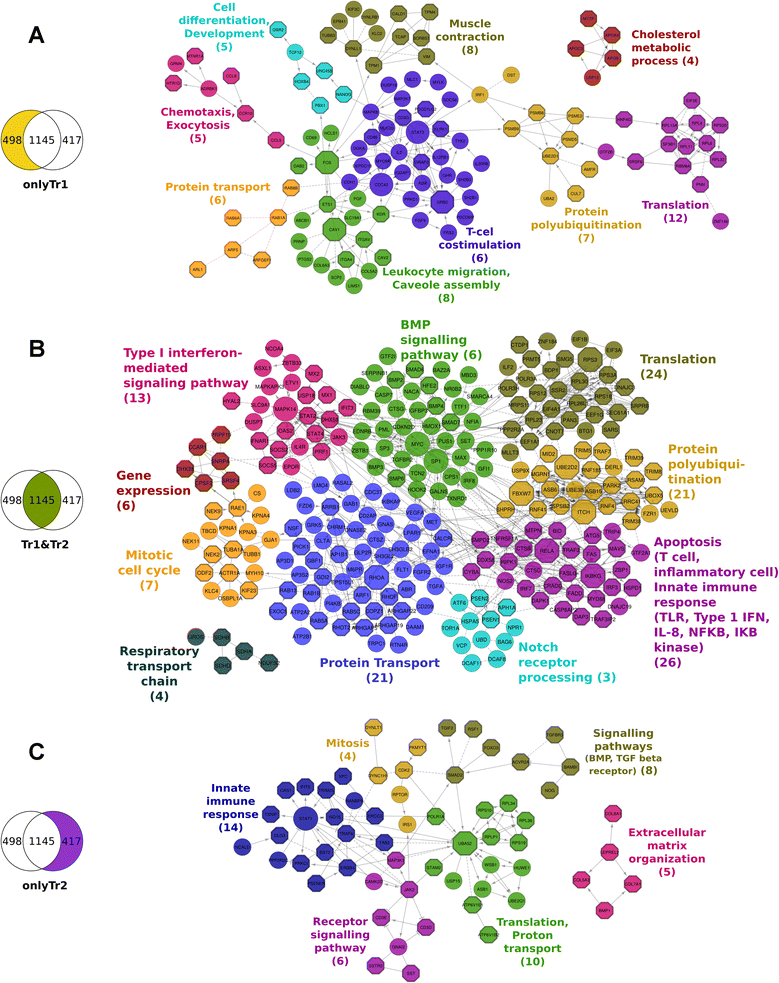
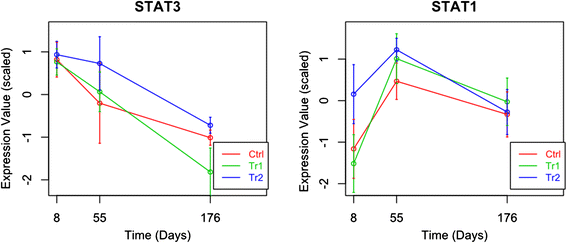
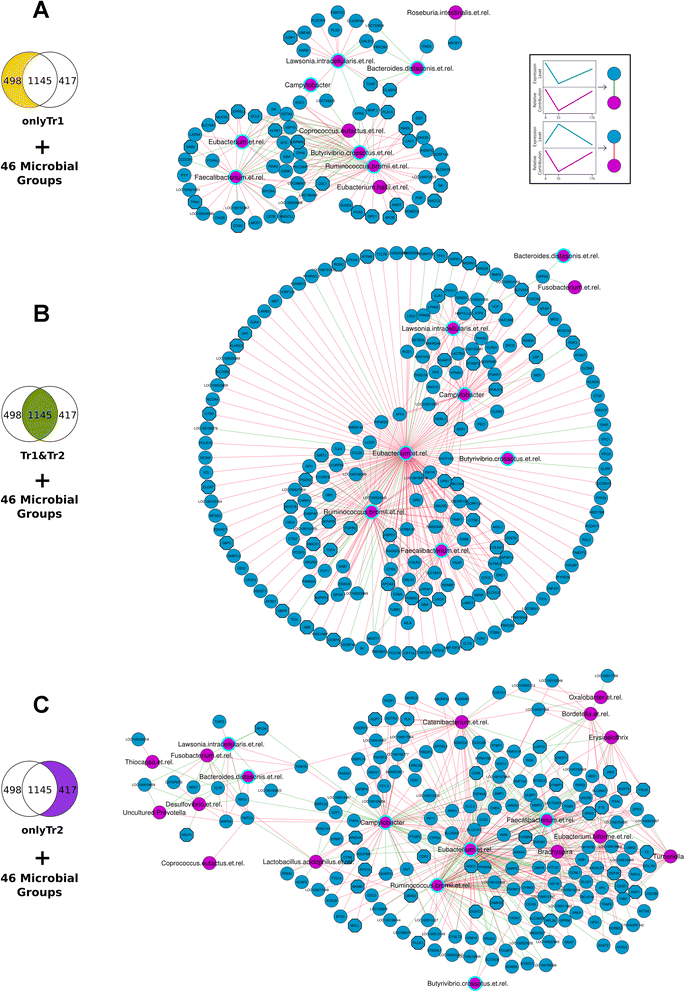
Results: The use of a new combination of data analysis tools allowed us to identify significant long-lasting differences in jejunal gene expression patterns resulting from the early life perturbations. In addition, we were able to identify potential key gene regulators (hubs) for these long-lasting effects. Furthermore, data integration also showed that there are a handful of bacterial groups that were associated with temporal changes in gene expression.
Conclusion: The applied systems-biology approach allowed us to take the first steps in unravelling biological processes involved in long lasting effects in the gut due to early life perturbations. The observed data are consistent with the hypothesis that these long lasting effects are due to differences in the programming of the gut immune system as induced by the temporary early life changes in the composition and/or diversity of microbiota in the gut.
Figures
References
-
- Foliaki S, Pearce N, Björkstén B, Mallol J, Montefort S, von Mutius E. Antibiotic use in infancy and symptoms of asthma, rhinoconjunctivitis, and eczema in children 6 and 7 years old: International Study of Asthma and Allergies in Childhood Phase III. J Allergy Clin Immunol. 2009;124:982–9. doi: 10.1016/j.jaci.2009.08.017. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

