Facilitating collaboration in rare genetic disorders through effective matchmaking in DECIPHER
- PMID: 26220709
- PMCID: PMC4832335
- DOI: 10.1002/humu.22842
Facilitating collaboration in rare genetic disorders through effective matchmaking in DECIPHER
Abstract
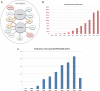
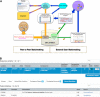
DECIPHER (https://decipher.sanger.ac.uk) is a web-based platform for secure deposition, analysis, and sharing of plausibly pathogenic genomic variants from well-phenotyped patients suffering from genetic disorders. DECIPHER aids clinical interpretation of these rare sequence and copy-number variants by providing tools for variant analysis and identification of other patients exhibiting similar genotype-phenotype characteristics. DECIPHER also provides mechanisms to encourage collaboration among a global community of clinical centers and researchers, as well as exchange of information between clinicians and researchers within a consortium, to accelerate discovery and diagnosis. DECIPHER has contributed to matchmaking efforts by enabling the global clinical genetics community to identify many previously undiagnosed syndromes and new disease genes, and has facilitated the publication of over 700 peer-reviewed scientific publications since 2004. At the time of writing, DECIPHER contains anonymized data from ∼250 registered centers on more than 51,500 patients (∼18000 patients with consent for data sharing and ∼25000 anonymized records shared privately). In this paper, we describe salient features of the platform, with special emphasis on the tools and processes that aid interpretation, sharing, and effective matchmaking with other data held in the database and that make DECIPHER an invaluable clinical and research resource.
Keywords: MatchMaker Exchange; genetic disorders; genotype-phenotype correlation; rare diseases.
© 2015 The Authors. **Human Mutation published by Wiley Periodicals, Inc.
Figures

References
-
- Barge‐Schaapveld DQCM, Maas SM, Polstra A, Knegt LC, Hennekam RC. 2011. The atypical 16p11.2 deletion: a not so atypical microdeletion syndrome? Am J Med Genet A 155A:1066–1072. - PubMed
-
- Boulding H, Webber C. 2012. Large‐scale objective association of mouse phenotypes with human symptoms through structural variation identified in patients with developmental disorders. Hum Mutat 33:874–883. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

