GeneMatcher: a matching tool for connecting investigators with an interest in the same gene
- PMID: 26220891
- PMCID: PMC4833888
- DOI: 10.1002/humu.22844
GeneMatcher: a matching tool for connecting investigators with an interest in the same gene
Abstract
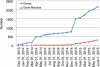
Here, we describe an overview and update on GeneMatcher (http://www.genematcher.org), a freely accessible Web-based tool developed as part of the Baylor-Hopkins Center for Mendelian Genomics. We created GeneMatcher with the goal of identifying additional individuals with rare phenotypes who had variants in the same candidate disease gene. We also wanted to facilitate connections to basic scientists working on orthologous genes in model systems with the goal of connecting their work to human Mendelian phenotypes. Meeting these goals will enhance the identification of novel Mendelian genes. Launched in September, 2013, GeneMatcher now has 2,178 candidate genes from 486 submitters spread across 38 countries entered in the database (June 1, 2015). GeneMatcher is also part of the Matchmaker Exchange (http://matchmakerexchange.org/) with an Application Programing Interface enabling submitters to query other databases of genetic variants and phenotypes without having to create accounts and data entries in multiple systems.
Keywords: Matchmaker Exchange; Mendelian disease; next-generation sequencing; variant analysis; whole exome sequencing; whole genome sequencing.
© 2015 WILEY PERIODICALS, INC.
Figures

References
-
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GCM, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(Database issue):D966–D974. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

