Association between α-synuclein blood transcripts and early, neuroimaging-supported Parkinson's disease
- PMID: 26220939
- PMCID: PMC4643625
- DOI: 10.1093/brain/awv202
Association between α-synuclein blood transcripts and early, neuroimaging-supported Parkinson's disease
Abstract
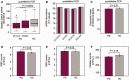
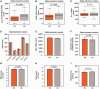
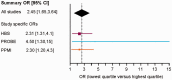
There are no cures for neurodegenerative diseases and this is partially due to the difficulty of monitoring pathogenic molecules in patients during life. The Parkinson's disease gene α-synuclein (SNCA) is selectively expressed in blood cells and neurons. Here we show that SNCA transcripts in circulating blood cells are paradoxically reduced in early stage, untreated and dopamine transporter neuroimaging-supported Parkinson's disease in three independent regional, national, and international populations representing 500 cases and 363 controls and on three analogue and digital platforms with P < 0.0001 in meta-analysis. Individuals with SNCA transcripts in the lowest quartile of counts had an odds ratio for Parkinson's disease of 2.45 compared to individuals in the highest quartile. Disease-relevant transcript isoforms were low even near disease onset. Importantly, low SNCA transcript abundance predicted cognitive decline in patients with Parkinson's disease during up to 5 years of longitudinal follow-up. This study reveals a consistent association of reduced SNCA transcripts in accessible peripheral blood and early-stage Parkinson's disease in 863 participants and suggests a clinical role as potential predictor of cognitive decline. Moreover, the three independent biobank cohorts provide a generally useful platform for rapidly validating any biological marker of this common disease.
Keywords: biobank; biomarker; cognitive decline; gene expression; mitochondria; α-synuclein.
© The Author (2015). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Comment in
-
A new biomarker for Parkinson's disease?Mov Disord. 2016 Jan;31(1):42. doi: 10.1002/mds.26448. Epub 2015 Dec 21. Mov Disord. 2016. PMID: 26688186 No abstract available.
References
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol 2009; 19: 465–74. - PubMed
-
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet 2003; 35: 292–3. - PubMed
-
- Braak H, Del Tredici K, Bratzke H, Hamm-Clement J, Sandmann-Keil D, Rub U. Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson's disease (preclinical and clinical stages). J Neurol 2002; 249 (Suppl 3): III/1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

