The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains
- PMID: 26220994
- PMCID: PMC4526735
- DOI: 10.1101/gad.267583.115
The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains
Abstract
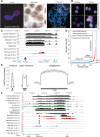
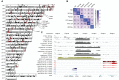
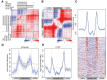
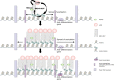
NUT midline carcinoma (NMC), a subtype of squamous cell cancer, is one of the most aggressive human solid malignancies known. NMC is driven by the creation of a translocation oncoprotein, BRD4-NUT, which blocks differentiation and drives growth of NMC cells. BRD4-NUT forms distinctive nuclear foci in patient tumors, which we found correlate with ∼100 unprecedented, hyperacetylated expanses of chromatin that reach up to 2 Mb in size. These "megadomains" appear to be the result of aberrant, feed-forward loops of acetylation and binding of acetylated histones that drive transcription of underlying DNA in NMC patient cells and naïve cells induced to express BRD4-NUT. Megadomain locations are typically cell lineage-specific; however, the cMYC and TP63 regions are targeted in all NMCs tested and play functional roles in tumor growth. Megadomains appear to originate from select pre-existing enhancers that progressively broaden but are ultimately delimited by topologically associating domain (TAD) boundaries. Therefore, our findings establish a basis for understanding the powerful role played by large-scale chromatin organization in normal and aberrant lineage-specific gene transcription.
Keywords: BRD4; chromatin hyperacetylation; topological domains.
© 2015 Alekseyenko et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Comment in
-
Epigenetics: Chromatin reorganization on a 'mega' scale.Nat Rev Cancer. 2015 Sep;15(9):512-3. doi: 10.1038/nrc4004. Epub 2015 Aug 13. Nat Rev Cancer. 2015. PMID: 26268835 No abstract available.
-
Epigenetics: Chromatin reorganization on a 'mega' scale.Nat Rev Genet. 2015 Sep;16(9):498-9. doi: 10.1038/nrg4000. Nat Rev Genet. 2015. PMID: 26281780 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
