Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis
- PMID: 26221021
- PMCID: PMC4538671
- DOI: 10.1073/pnas.1513033112
Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis
Abstract
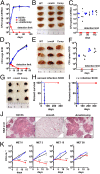
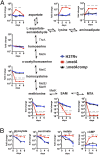
Multidrug resistance, strong side effects, and compliance problems in TB chemotherapy mandate new ways to kill Mycobacterium tuberculosis (Mtb). Here we show that deletion of the gene encoding homoserine transacetylase (metA) inactivates methionine and S-adenosylmethionine (SAM) biosynthesis in Mtb and renders this pathogen exquisitely sensitive to killing in immunocompetent or immunocompromised mice, leading to rapid clearance from host tissues. Mtb ΔmetA is unable to proliferate in primary human macrophages, and in vitro starvation leads to extraordinarily rapid killing with no appearance of suppressor mutants. Cell death of Mtb ΔmetA is faster than that of other auxotrophic mutants (i.e., tryptophan, pantothenate, leucine, biotin), suggesting a particularly potent mechanism of killing. Time-course metabolomics showed complete depletion of intracellular methionine and SAM. SAM depletion was consistent with a significant decrease in methylation at the DNA level (measured by single-molecule real-time sequencing) and with the induction of several essential methyltransferases involved in biotin and menaquinone biosynthesis, both of which are vital biological processes and validated targets of antimycobacterial drugs. Mtb ΔmetA could be partially rescued by biotin supplementation, confirming a multitarget cell death mechanism. The work presented here uncovers a previously unidentified vulnerability of Mtb-the incapacity to scavenge intermediates of SAM and methionine biosynthesis from the host. This vulnerability unveils an entirely new drug target space with the promise of rapid killing of the tubercle bacillus by a new mechanism of action.
Keywords: amino acid biosynthesis; bactericidal auxotrophy; host–pathogen interaction; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sévin DC, Kuehne A, Zamboni N, Sauer U. Biological insights through nontargeted metabolomics. Curr Opin Biotechnol. 2014;34C:1–8. - PubMed
-
- Ehrt S, Rhee K. Mycobacterium tuberculosis metabolism and host interaction: Mysteries and paradoxes. Curr Top Microbiol Immunol. 2013;374:163–188. - PubMed
-
- Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8(6):401–412. - PubMed
-
- Horn M, et al. Illuminating the evolutionary history of chlamydiae. Science. 2004;304(5671):728–730. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

