Knockout of SRC-1 and SRC-3 in Mice Decreases Cardiomyocyte Proliferation and Causes a Noncompaction Cardiomyopathy Phenotype
- PMID: 26221073
- PMCID: PMC4515817
- DOI: 10.7150/ijbs.12408
Knockout of SRC-1 and SRC-3 in Mice Decreases Cardiomyocyte Proliferation and Causes a Noncompaction Cardiomyopathy Phenotype
Abstract
Noncompaction cardiomyopathy (NCC) is a congenital heart disease that causes ventricular dysfunction and high mortality rate in children. The mechanisms responsible for NCC are still unknown. The steroid receptor coactivator-1 (SRC-1) and SRC-3 are transcriptional coactivators for nuclear hormone receptors and certain other transcription factors that regulate many genes in development and organ function. However, the roles of SRC-1/3 in heart morphogenesis, function and NCC occurrence are unknown. This study aims to examine the spatial and temporal expression patterns of SRC-1/3 in the heart and investigate the specific roles of SRC-1/3 in heart development, function and NCC occurrence. Immunochemical analysis detected SRC-1/3 expressions in the proliferating cardiomyocytes of mouse heart at prenatal and neonatal stages, while these expressions disappeared within two weeks after birth. Through generating and characterizing mouse lines with global or cardiomyocyte-specific knockouts of SRC-1/3, we found ablation of SRC-1/3 in the myocardial lineage resulted in prominent trabeculae, deep intertrabecular recesses and thin ventricular wall and septum. These developmental defects caused a failure of trabecular compaction, decreased internal ventricular dimension, reduced cardiac ejection fraction and output and led to a high rate of postnatal mortality. Collectively, these structural and functional abnormalities closely simulate the phenotype of NCC patients. Further molecular analysis of cardiomyocytes in vivo and in vitro revealed that SRC-1/3 directly up-regulate cyclin E2, cyclin B1 and myocardin to promote cardiomyocyte proliferation and differentiation. In conclusion, SRC-1/3 are required for cardiomyocyte proliferation and differentiation at earlier developmental stages, and their dysfunction causes NCC-like abnormalities in the hearts of newborn and adult mice.
Keywords: cardiac output; heart diseases; knockout mice; myocyte proliferation; nuclear receptor coactivator.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

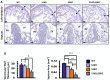
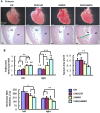

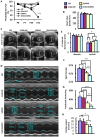
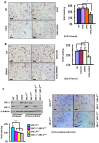

References
-
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900. - PubMed
-
- Marelli AJ, Mackie AS, Ionescu-Ittu R. et al. Congenital heart disease in the general population: changing prevalence and age distribution. Circulation. 2007;115:163–72. - PubMed
-
- Bruneau BG. The developmental genetics of congenital heart disease. nature. 2008;451:943–8. - PubMed
-
- Weiford BC, Subbarao VD, Mulhern KM. Noncompaction of the ventricular myocardium. Circulation. 2004;109:2965–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

