Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas
- PMID: 26221190
- PMCID: PMC4517542
- DOI: 10.1186/s13148-015-0091-3
Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas
Abstract
Background: Recent developments in genomic sequencing have advanced our understanding of the mutations underlying human malignancy. Melanoma is a prototype of an aggressive, genetically heterogeneous cancer notorious for its biologic plasticity and predilection towards developing resistance to targeted therapies. Evidence is rapidly accumulating that dysregulated epigenetic mechanisms (DNA methylation/demethylation, histone modification, non-coding RNAs) may play a central role in the pathogenesis of melanoma. Therefore, we sought to characterize the frequency and nature of mutations in epigenetic regulators in clinical, treatment-naïve, patient melanoma specimens obtained from one academic institution.
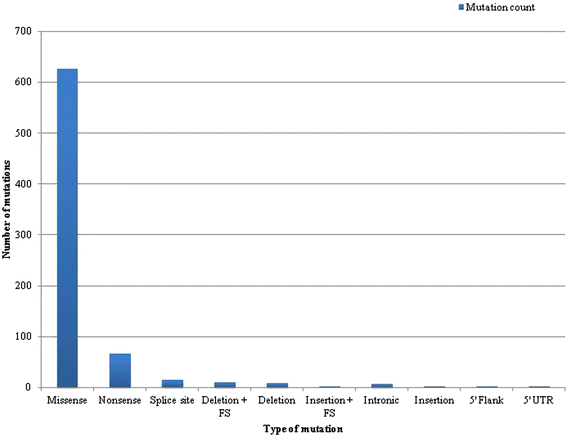
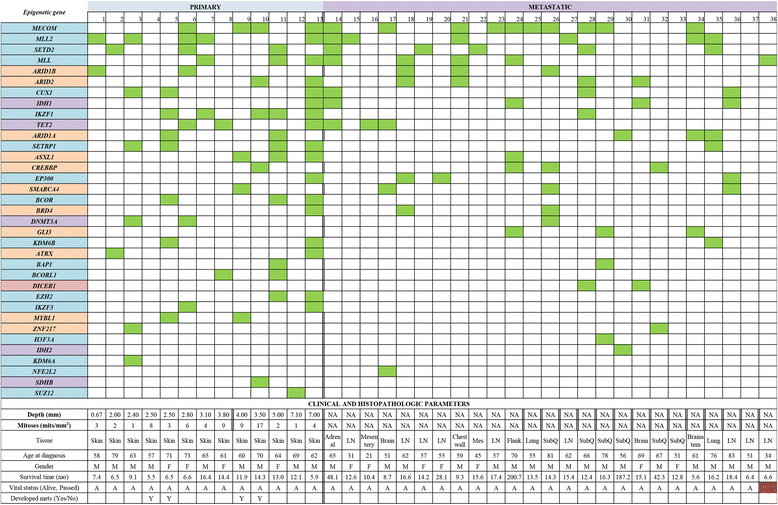
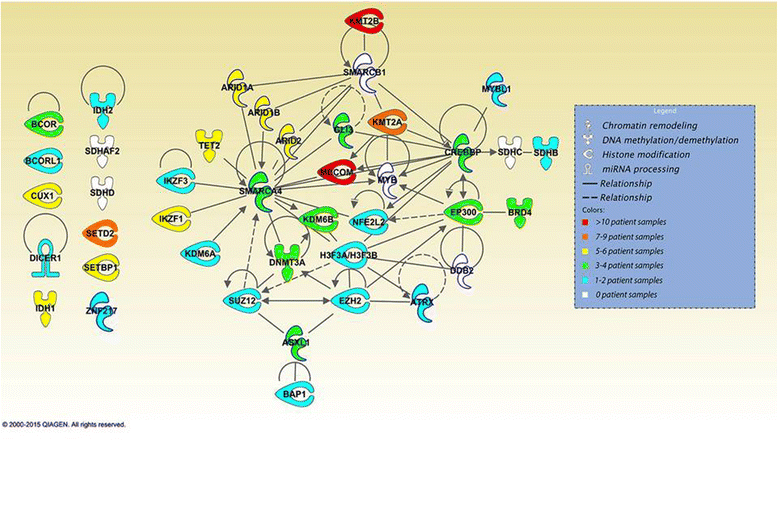
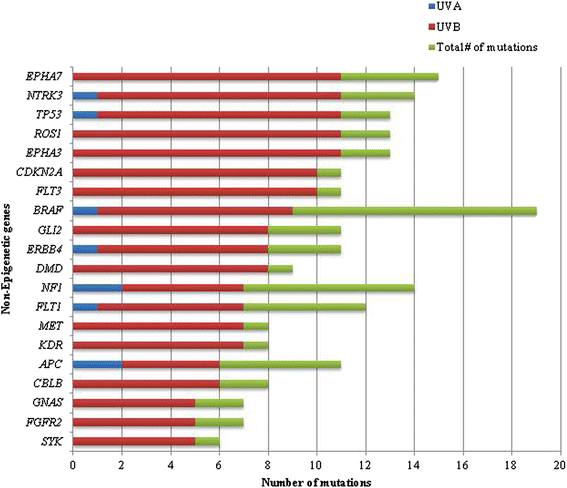
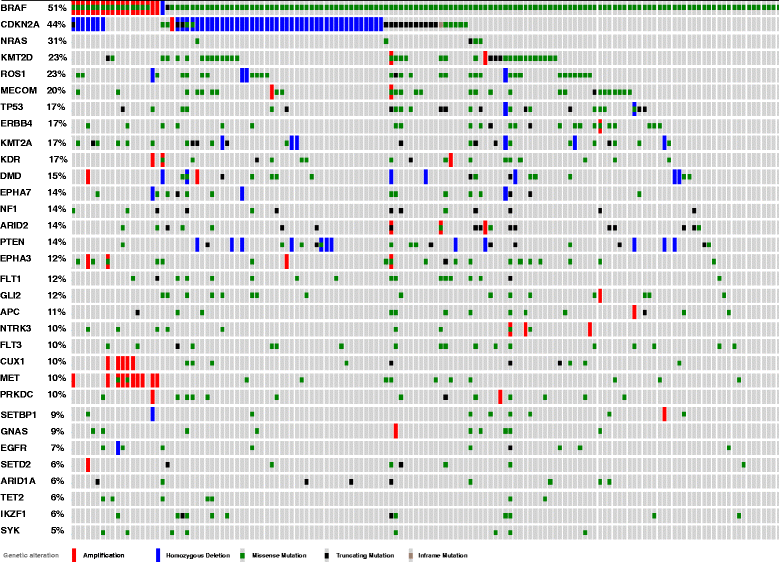
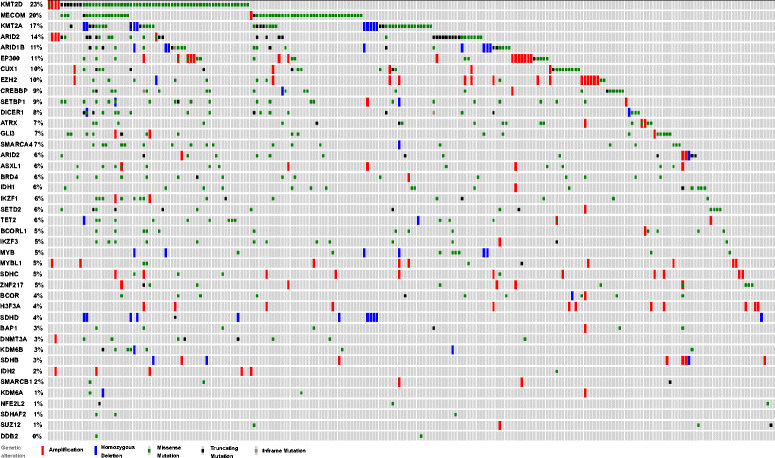
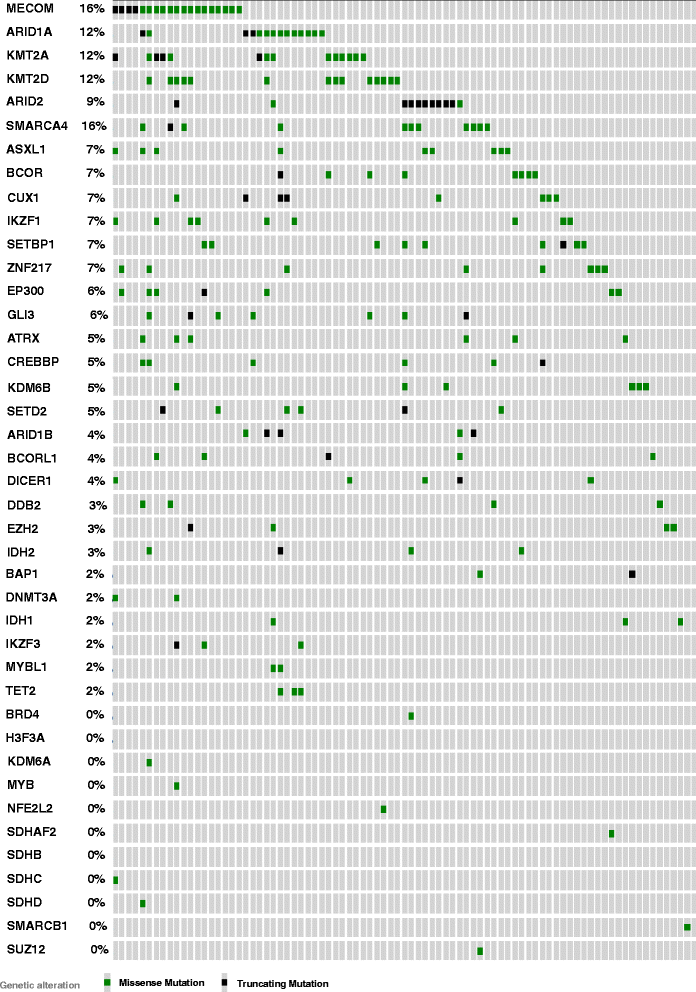
Results: Targeted next-generation sequencing for 275 known and investigative cancer genes (of which 41 genes, or 14.9 %, encoded an epigenetic regulator) of 38 treatment-naïve patient melanoma samples revealed that 22.3 % (165 of 740) of all non-silent mutations affected an epigenetic regulator. The most frequently mutated genes were BRAF, MECOM, NRAS, TP53, MLL2, and CDKN2A. Of the 40 most commonly mutated genes, 12 (30.0 %) encoded epigenetic regulators, including genes encoding enzymes involved in histone modification (MECOM, MLL2, SETD2), chromatin remodeling (ARID1B, ARID2), and DNA methylation and demethylation (TET2, IDH1). Among the 38 patient melanoma samples, 35 (92.1 %) harbored at least one mutation in an epigenetic regulator. The genes with the highest number of total UVB-signature mutations encoded epigenetic regulators, including MLL2 (100 %, 16 of 16) and MECOM (82.6 %, 19 of 23). Moreover, on average, epigenetic genes harbored a significantly greater number of UVB-signature mutations per gene than non-epigenetic genes (3.7 versus 2.4, respectively; p = 0.01). Bioinformatics analysis of The Cancer Genome Atlas (TCGA) melanoma mutation dataset also revealed a frequency of mutations in the 41 epigenetic genes comparable to that found within our cohort of patient melanoma samples.
Conclusions: Our study identified a high prevalence of somatic mutations in genes encoding epigenetic regulators, including those involved in DNA demethylation, histone modification, chromatin remodeling, and microRNA processing. Moreover, UVB-signature mutations were found more commonly among epigenetic genes than in non-epigenetic genes. Taken together, these findings further implicate epigenetic mechanisms, particularly those involving the chromatin-remodeling enzyme MECOM/EVI1 and histone-modifying enzyme MLL2, in the pathobiology of melanoma.
Keywords: 5-hydroxymethylcytosine; DNA demethylation; Epigenetics; Isocitrate dehydrogenase 2 (IDH2); MECOM (MDS1 and EV1 complex locus); MLL2; Melanoma; Next-generation sequencing (NGS); Ten-eleven translocation (TET).
Figures





References
-
- Lian CG, Mihm MC, Pierard G, Tommasino M. Skin Cancer. In: Stewart BW, Wild CP, editors. World Cancer Report 2014. Lyon, France: International Agency for Research on Cancer; 2014.
-
- Garraway LA, Chin L. Chapter 118: molecular biology of cutaneous melanoma. In: Devita VT Jr, Lawrence TS, Rosenberg SA, editors. Cancer: Principles & Practice of Oncology, 9e. Philadelphia, PA: Lippincott Williams & Wilkins; 2011.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

