Genome sequence of Clostridium sporogenes DSM 795(T), an amino acid-degrading, nontoxic surrogate of neurotoxin-producing Clostridium botulinum
- PMID: 26221421
- PMCID: PMC4517662
- DOI: 10.1186/s40793-015-0016-y
Genome sequence of Clostridium sporogenes DSM 795(T), an amino acid-degrading, nontoxic surrogate of neurotoxin-producing Clostridium botulinum
Abstract
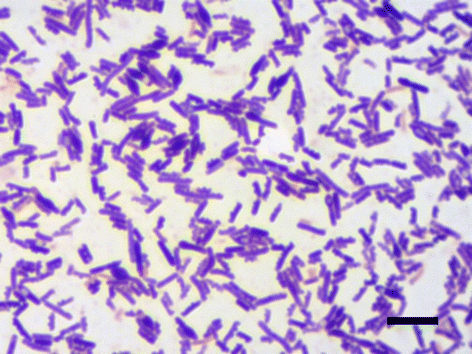
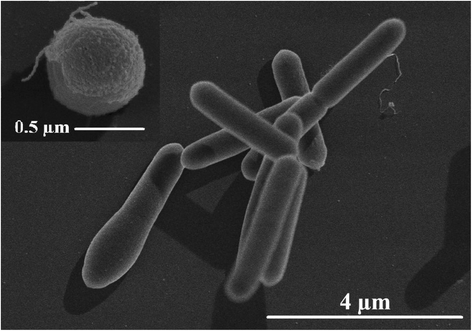
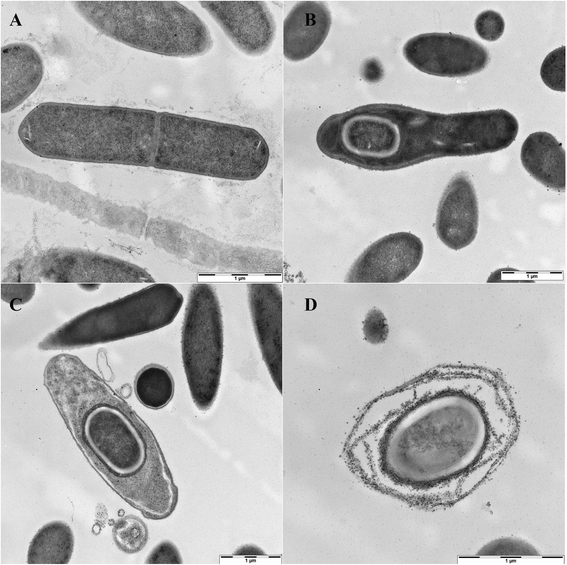
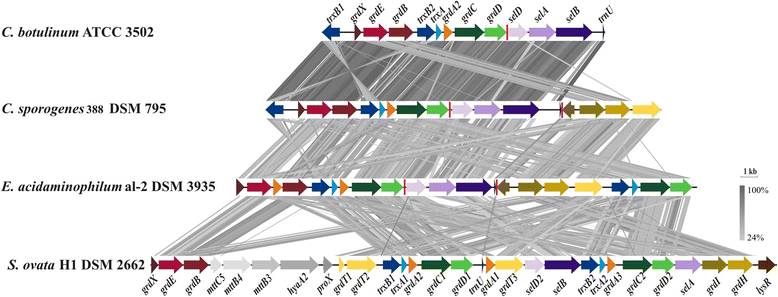
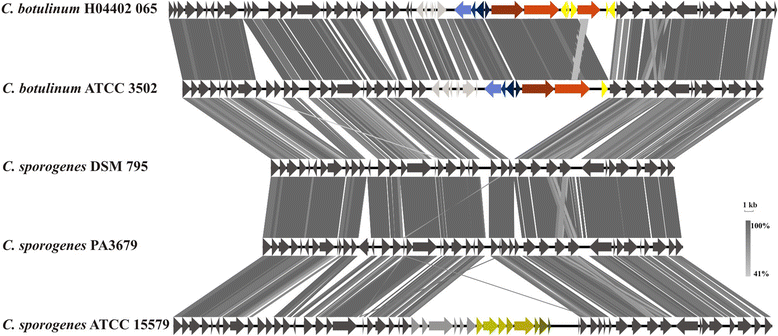
Clostridium sporogenes DSM 795 is the type strain of the species Clostridium sporogenes, first described by Metchnikoff in 1908. It is a Gram-positive, rod-shaped, anaerobic bacterium isolated from human faeces and belongs to the proteolytic branch of clostridia. C. sporogenes attracts special interest because of its potential use in a bacterial therapy for certain cancer types. Genome sequencing and annotation revealed several gene clusters coding for proteins involved in anaerobic degradation of amino acids, such as glycine and betaine via Stickland reaction. Genome comparison showed that C. sporogenes is closely related to C. botulinum. The genome of C. sporogenes DSM 795 consists of a circular chromosome of 4.1 Mb with an overall GC content of 27.81 mol% harboring 3,744 protein-coding genes, and 80 RNAs.
Keywords: Anaerobic; Butanol; C. botulinum; C. sporogenes; Gram-positive; Stickland reaction.
Figures

Similar articles
-
Implications of Genome-Based Discrimination between Clostridium botulinum Group I and Clostridium sporogenes Strains for Bacterial Taxonomy.Appl Environ Microbiol. 2015 Aug 15;81(16):5420-9. doi: 10.1128/AEM.01159-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048939 Free PMC article.
-
Differentiating Botulinum Neurotoxin-Producing Clostridia with a Simple, Multiplex PCR Assay.Appl Environ Microbiol. 2017 Aug 31;83(18):e00806-17. doi: 10.1128/AEM.00806-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28733282 Free PMC article.
-
Clostridium tepidum sp. nov., a close relative of Clostridium sporogenes and Clostridium botulinum Group I.Int J Syst Evol Microbiol. 2017 Jul;67(7):2317-2322. doi: 10.1099/ijsem.0.001948. Epub 2017 Jul 10. Int J Syst Evol Microbiol. 2017. PMID: 28693684
-
Clostridium sporogenes PA 3679 and its uses in the derivation of thermal processing schedules for low-acid shelf-stable foods and as a research model for proteolytic Clostridium botulinum.J Food Prot. 2012 Apr;75(4):779-92. doi: 10.4315/0362-028X.JFP-11-391. J Food Prot. 2012. PMID: 22488072 Review.
-
Clostridium botulinum: a bug with beauty and weapon.Crit Rev Microbiol. 2005;31(1):11-8. doi: 10.1080/10408410590912952. Crit Rev Microbiol. 2005. PMID: 15839401 Review.
Cited by
-
Molecular Keys to the Janthinobacterium and Duganella spp. Interaction with the Plant Pathogen Fusarium graminearum.Front Microbiol. 2016 Oct 26;7:1668. doi: 10.3389/fmicb.2016.01668. eCollection 2016. Front Microbiol. 2016. PMID: 27833590 Free PMC article.
-
Genomics of the Pathogenic Clostridia.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0033-2018. doi: 10.1128/microbiolspec.GPP3-0033-2018. Microbiol Spectr. 2019. PMID: 31215504 Free PMC article. Review.
-
SpoIVA is an essential morphogenetic protein for the formation of heat- and lysozyme-resistant spores in Clostridium sporogenes NBRC 14293.Front Microbiol. 2024 Apr 24;15:1338751. doi: 10.3389/fmicb.2024.1338751. eCollection 2024. Front Microbiol. 2024. PMID: 38721605 Free PMC article.
-
Tryptophan was metabolized into beneficial metabolites against coronary heart disease or prevented from producing harmful metabolites by the in vitro drug screening model based on Clostridium sporogenes.Front Microbiol. 2022 Nov 16;13:1013973. doi: 10.3389/fmicb.2022.1013973. eCollection 2022. Front Microbiol. 2022. PMID: 36466649 Free PMC article.
-
Culture-based and culture-independent approach for the study of the methanogens and obligate anaerobes from different landfill sites.Front Microbiol. 2024 Jan 29;14:1273037. doi: 10.3389/fmicb.2023.1273037. eCollection 2023. Front Microbiol. 2024. PMID: 38348306 Free PMC article.
References
-
- Metchnikoff E. Sur les microbes de la putréfacion intestinale. C R Hebd Seances Acad Sci. 1908;147:579–82.
-
- Metchnikoff E. Etude sur la flore intestinale. IV. Le Bacillus sporogenes. Annales de l’Institut Pasteur. 1908;22:942–6.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

