Genome Wide Association Study for Predictors of Progression Free Survival in Patients on Capecitabine, Oxaliplatin, Bevacizumab and Cetuximab in First-Line Therapy of Metastatic Colorectal Cancer
- PMID: 26222057
- PMCID: PMC4519298
- DOI: 10.1371/journal.pone.0131091
Genome Wide Association Study for Predictors of Progression Free Survival in Patients on Capecitabine, Oxaliplatin, Bevacizumab and Cetuximab in First-Line Therapy of Metastatic Colorectal Cancer
Abstract
Purpose: Despite expanding options for systemic treatment, survival for metastatic colorectal cancer (mCRC) remains limited and individual response is difficult to predict. In search of pre-treatment predictors, pharmacogenetic research has mainly used a candidate gene approach. Genome wide association (GWA) studies offer the benefit of simultaneously analyzing a large number of SNPs, in both known and still unidentified functional regions. Using a GWA approach, we searched for genetic markers affecting progression free survival (PFS) in mCRC patients treated with first-line capecitabine, oxaliplatin and bevacizumab (CAPOX-B), with or without cetuximab.
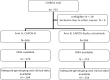
Patients and methods: 755 patients were included in the CAIRO2-trial, a multicenter phase III trial, randomizing between first-line treatment with CAPOX-B versus CAPOX-B plus cetuximab. Germline DNA and complete clinical information was available from 553 patients and genome wide genotyping was performed, using Illumina's OmniExpress beadchip arrays, with 647,550 markers passing all quality checks. Another 2,202,473 markers were imputated by using HapMap2. Association with PFS was analysed using a Cox proportional hazards model.
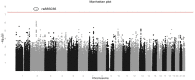
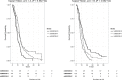
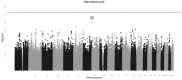
Results: One marker, rs885036, associated significantly with PFS (P = 2.17x10(-8)) showing opposite effects on PFS depending on treatment arm. The minor allele was associated with increased PFS in patients receiving cetuximab. A cluster of markers located on chromosome 8 associated with PFS, irrespective of treatment arm (P-values of 2.30x10(-7) to 1.04x10(-6)).
Conclusion: This is the first GWA study to find SNPs affecting PFS in mCRC patients treated with CAPOX-B, either with or without cetuximab. Rs885036 is a potential predictive marker for cetuximab efficacy. These markers need to be validated in independent treatment cohorts.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

