NRF2 Signaling Negatively Regulates Phorbol-12-Myristate-13-Acetate (PMA)-Induced Differentiation of Human Monocytic U937 Cells into Pro-Inflammatory Macrophages
- PMID: 26222138
- PMCID: PMC4519053
- DOI: 10.1371/journal.pone.0134235
NRF2 Signaling Negatively Regulates Phorbol-12-Myristate-13-Acetate (PMA)-Induced Differentiation of Human Monocytic U937 Cells into Pro-Inflammatory Macrophages
Abstract
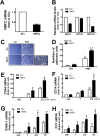
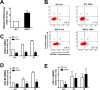
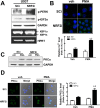
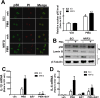
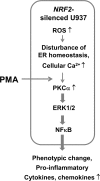
Blood monocytes are recruited to injured tissue sites and differentiate into macrophages, which protect against pathogens and repair damaged tissues. Reactive oxygen species (ROS) are known to be an important contributor to monocytes' differentiation and macrophages' function. NF-E2-related factor 2 (NRF2), a transcription factor regulating cellular redox homeostasis, is known to be a critical modulator of inflammatory responses. We herein investigated the role of NRF2 in macrophage differentiation using the human monocytic U937 cell line and phorbol-12-myristate-13-acetate (PMA). In U937 cells with NRF2 silencing, PMA-stimulated cell adherence was significantly facilitated when compared to control U937 cells. Both transcript and protein levels for pro-inflammatory cytokines, including interleukine-1β (IL-1β), IL-6, and tumor necrosis factor-α (TNFα) were highly elevated in PMA-stimulated NRF2-silenced U937 compared to the control. In addition, PMA-inducible secretion of monocyte chemotactic protein 1 (MCP-1) was significantly high in NRF2-silenced U937. As an underlying mechanism, we showed that NRF2-knockdown U937 retained high levels of cellular ROS and endoplasmic reticulum (ER) stress markers expression; and subsequently, PMA-stimulated levels of Ca2+ and PKCα were greater in NRF2-knockdown U937 cells, which caused enhanced nuclear accumulation of nuclear factor-ҡB (NFҡB) p50 and extracellular signal-regulated kinase (ERK)-1/2 phosphorylation. Whereas the treatment of NRF2-silenced U937 cells with pharmacological inhibitors of NFҡB or ERK1/2 largely blocked PMA-induced IL-1β and IL-6 expression, indicating that these pathways are associated with cell differentiation. Taken together, our results suggest that the NRF2 system functions to suppress PMA-stimulated U937 cell differentiation into pro-inflammatory macrophages and provide evidence that the ROS-PKCα-ERK-NFҡB axis is involved in PMA-facilitated differentiation of NRF2-silenced U937 cells.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

