MStern Blotting-High Throughput Polyvinylidene Fluoride (PVDF) Membrane-Based Proteomic Sample Preparation for 96-Well Plates
- PMID: 26223766
- PMCID: PMC4597154
- DOI: 10.1074/mcp.O115.049650
MStern Blotting-High Throughput Polyvinylidene Fluoride (PVDF) Membrane-Based Proteomic Sample Preparation for 96-Well Plates
Abstract
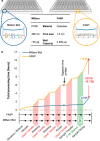
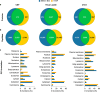
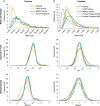
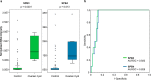
We describe a 96-well plate compatible membrane-based proteomic sample processing method, which enables the complete processing of 96 samples (or multiples thereof) within a single workday. This method uses a large-pore hydrophobic PVDF membrane that efficiently adsorbs proteins, resulting in fast liquid transfer through the membrane and significantly reduced sample processing times. Low liquid transfer speeds have prevented the useful 96-well plate implementation of FASP as a widely used membrane-based proteomic sample processing method. We validated our approach on whole-cell lysate and urine and cerebrospinal fluid as clinically relevant body fluids. Without compromising peptide and protein identification, our method uses a vacuum manifold and circumvents the need for digest desalting, making our processing method compatible with standard liquid handling robots. In summary, our new method maintains the strengths of FASP and simultaneously overcomes one of the major limitations of FASP without compromising protein identification and quantification.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Manza L. L., Stamer S. L., Ham A. J., Codreanu S. G., Liebler D. C. (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 - PubMed
-
- Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nature Meth. 6, 359–362 - PubMed
-
- Switzar L., van Angeren J., Pinkse M., Kool J., Niessen W. M. (2013) A high-throughput sample preparation method for cellular proteomics using 96-well filter plates. Proteomics 13, 2980–2983 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

