Molecular characterization of ring chromosome 18 by low-coverage next generation sequencing
- PMID: 26224010
- PMCID: PMC4557216
- DOI: 10.1186/s12881-015-0206-x
Molecular characterization of ring chromosome 18 by low-coverage next generation sequencing
Abstract
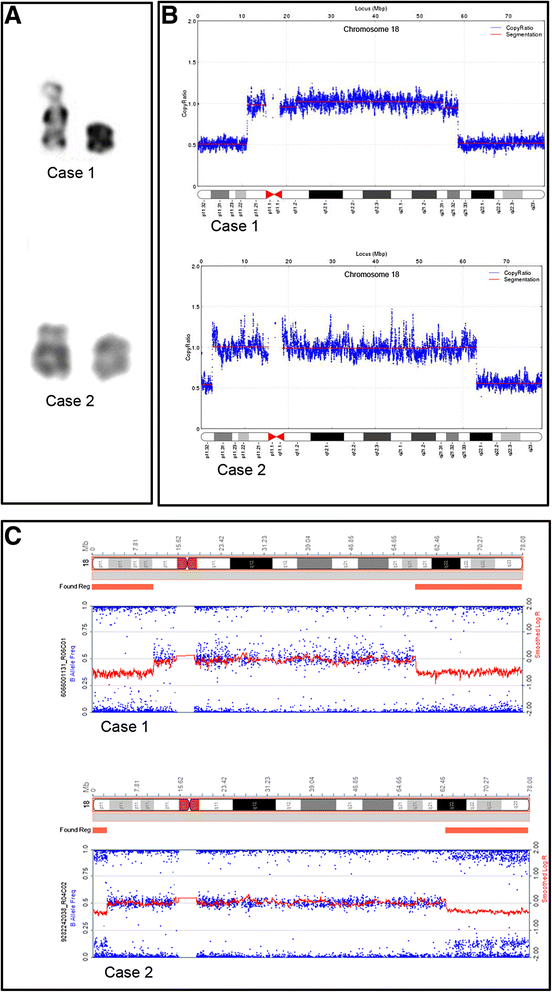
Background: Ring chromosomes are one category of structurally abnormal chromosomes that can lead to severe growth retardation and other clinical defects. Traditionally, their diagnosis and characterization has largely relied on conventional cytogenetics and fluorescence in situ hybridization, array-based comparative genomic hybridization and single nucleotide polymorphism array-based comparative genomic hybridization. However, these methods are ineffectively at characterizing the ring chromosome structure and only offer a low resolution mapping of breakpoints. Here, we applied whole-genome low-coverage paired-end next generation sequencing (NGS) to two suspected cases of ring chromosome 18 (r(18)) and characterized the ring structure including the chromosome dosage changes and the breakpoint junction.
Methods: The breakpoints and chromosome copy number variations (CNVs) of r(18) were characterized by whole-genome low-coverage paired-end NGS. We confirmed the dosage change by single nucleotide polymorphisms array, and validated the junction site regions using PCR followed by Sanger sequencing.
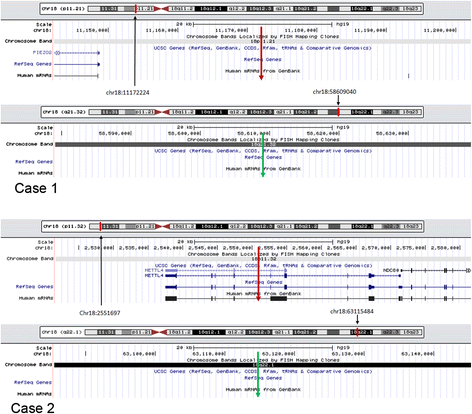
Results: We successfully and fully characterized the r(18) in two cases by NGS. We mapped the breakpoints with a high resolution and identified all CNVs in both cases. We analyzed the breakpoint regions and discovered two breakpoints located within repetitive sequence regions, and two near the repetitive sequence regions. One of the breakpoints in case 2 was located within the gene METTL4, while the other breakpoints were intergenic.
Conclusions: We demonstrated that whole-genome low-coverage paired-end NGS can be used directly to map breakpoints with a high molecular resolution and detect all CNVs on r(18). This approach will provide new insights into the genotype-phenotype correlations on r(18) and the underlying mechanism of ring chromosomes formation. Our results also demonstrate that this can be a powerful approach for the diagnosis and characterization of ring chromosomes in the clinic.
Figures

References
-
- Stankiewicz P, Brozek I, Helias-Rodzewicz Z, Wierzba J, Pilch J, Bocian E, Balcerska A, Wozniak A, Kardas I, Wirth J, et al. Clinical and molecular-cytogenetic studies in seven patients with ring chromosome 18. Am J Med Genet. 2001;101(3):226–239. doi: 10.1002/1096-8628(20010701)101:3<226::AID-AJMG1349>3.0.CO;2-#. - DOI - PubMed
-
- Carter E, Heard P, Hasi M, Soileau B, Sebold C, Hale DE, Cody JD.Ring 18 molecular assessment and clinical consequences. American journal of medical genetics Part A. 2015;167A(1):54-63. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

