Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription
- PMID: 26224331
- PMCID: PMC4519778
- DOI: 10.1038/srep12524
Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription
Abstract
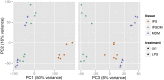
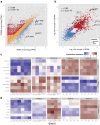
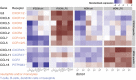
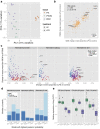
Macrophages differentiated from human induced pluripotent stem cells (IPSDMs) are a potentially valuable new tool for linking genotype to phenotype in functional studies. However, at a genome-wide level these cells have remained largely uncharacterised. Here, we compared the transcriptomes of naïve and lipopolysaccharide (LPS) stimulated monocyte-derived macrophages (MDMs) and IPSDMs using RNA-Seq. The IPSDM and MDM transcriptomes were broadly similar and exhibited a highly conserved response to LPS. However, there were also significant differences in the expression of genes associated with antigen presentation and tissue remodelling. Furthermore, genes coding for multiple chemokines involved in neutrophil recruitment were more highly expressed in IPSDMs upon LPS stimulation. Additionally, analysing individual transcript expression identified hundreds of genes undergoing alternative promoter and 3' untranslated region usage following LPS treatment representing a previously under-appreciated level of regulation in the LPS response.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

