ETS-related transcription factors ETV4 and ETV5 are involved in proliferation and induction of differentiation-associated genes in embryonic stem (ES) cells
- PMID: 26224636
- PMCID: PMC4566222
- DOI: 10.1074/jbc.M115.675595
ETS-related transcription factors ETV4 and ETV5 are involved in proliferation and induction of differentiation-associated genes in embryonic stem (ES) cells
Abstract
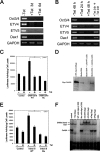
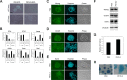
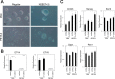
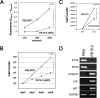
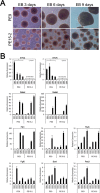
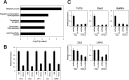
The pluripotency and self-renewal capacity of embryonic stem (ES) cells is regulated by several transcription factors. Here, we show that the ETS-related transcription factors Etv4 and Etv5 (Etv4/5) are specifically expressed in undifferentiated ES cells, and suppression of Oct3/4 results in down-regulation of Etv4/5. Simultaneous deletion of Etv4 and Etv5 (Etv4/5 double knock-out (dKO)) in ES cells resulted in a flat, epithelial cell-like appearance, whereas the morphology changed into compact colonies in a 2i medium (containing two inhibitors for GSK3 and MEK/ERK). Expression levels of self-renewal marker genes, including Oct3/4 and Nanog, were similar between wild-type and dKO ES cells, whereas proliferation of Etv4/5 dKO ES cells was decreased with overexpression of cyclin-dependent kinase inhibitors (p16/p19, p15, and p57). A differentiation assay revealed that the embryoid bodies derived from Etv4/5 dKO ES cells were smaller than the control, and expression of ectoderm marker genes, including Fgf5, Sox1, and Pax3, was not induced in dKO-derived embryoid bodies. Microarray analysis demonstrated that stem cell-related genes, including Tcf15, Gbx2, Lrh1, Zic3, and Baf60c, were significantly repressed in Etv4/5 dKO ES cells. The artificial expression of Etv4 and/or Etv5 in Etv4/5 dKO ES cells induced re-expression of Tcf15 and Gbx2. These results indicate that Etv4 and Etv5, potentially through regulation of Gbx2 and Tcf15, are involved in the ES cell proliferation and induction of differentiation-associated genes in ES cells.
Keywords: cell proliferation; differentiation; embryonic stem cell; gene regulation; oncogene; transcription factor.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 - PubMed
-
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988) Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 - PubMed
-
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 - PubMed
-
- Niwa H., Ogawa K., Shimosato D., Adachi K. (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

