BioModels: Content, Features, Functionality, and Use
- PMID: 26225232
- PMCID: PMC4360671
- DOI: 10.1002/psp4.3
BioModels: Content, Features, Functionality, and Use
Abstract
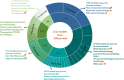
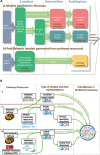
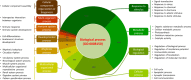
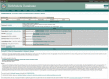
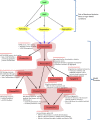
BioModels is a reference repository hosting mathematical models that describe the dynamic interactions of biological components at various scales. The resource provides access to over 1,200 models described in literature and over 140,000 models automatically generated from pathway resources. Most model components are cross-linked with external resources to facilitate interoperability. A large proportion of models are manually curated to ensure reproducibility of simulation results. This tutorial presents BioModels' content, features, functionality, and usage.
Figures
References
-
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. - PubMed
-
- Juty N, Le Novère N. Laibe C. Identifiers.org and MIRIAM registry: community resources to provide persistent identification. Nucleic Acids Res. 2012;40:D580–D586. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

