Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort
- PMID: 26226137
- PMCID: PMC4733433
- DOI: 10.1038/gim.2015.89
Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort
Erratum in
-
CORRIGENDUM: Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort.Genet Med. 2016 Aug;18(8):859. doi: 10.1038/gim.2016.84. Genet Med. 2016. PMID: 27494218 No abstract available.
Abstract
Purpose: Autosomal recessive nonsyndromic deafness (ARNSD) is characterized by a high degree of genetic heterogeneity, with reported mutations in 58 different genes. This study was designed to detect deafness-causing variants in a multiethnic cohort with ARNSD by using whole-exome sequencing (WES).
Methods: After excluding mutations in the most common gene, GJB2, we performed WES in 160 multiplex families with ARNSD from Turkey, Iran, Mexico, Ecuador, and Puerto Rico to screen for mutations in all known ARNSD genes.
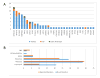
Results: We detected ARNSD-causing variants in 90 (56%) families, 54% of which had not been previously reported. Identified mutations were located in 31 known ARNSD genes. The most common genes with mutations were MYO15A (13%), MYO7A (11%), SLC26A4 (10%), TMPRSS3 (9%), TMC1 (8%), ILDR1 (6%), and CDH23 (4%). Nine mutations were detected in multiple families with shared haplotypes, suggesting founder effects.
Conclusion: We report on a large multiethnic cohort with ARNSD in which comprehensive analysis of all known ARNSD genes identifies causative DNA variants in 56% of the families. In the remaining families, WES allows us to search for causative variants in novel genes, thus improving our ability to explain the underlying etiology in more families.Genet Med 18 4, 364-371.
Conflict of interest statement
Figures


References
-
- Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

