Stochasticity and Spatial Interaction Govern Stem Cell Differentiation Dynamics
- PMID: 26227093
- PMCID: PMC4521170
- DOI: 10.1038/srep12617
Stochasticity and Spatial Interaction Govern Stem Cell Differentiation Dynamics
Abstract
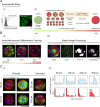
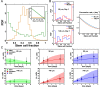
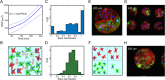
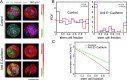
Stem cell differentiation underlies many fundamental processes such as development, tissue growth and regeneration, as well as disease progression. Understanding how stem cell differentiation is controlled in mixed cell populations is an important step in developing quantitative models of cell population dynamics. Here we focus on quantifying the role of cell-cell interactions in determining stem cell fate. Toward this, we monitor stem cell differentiation in adherent cultures on micropatterns and collect statistical cell fate data. Results show high cell fate variability and a bimodal probability distribution of stem cell fraction on small (80-140 μm diameter) micropatterns. On larger (225-500 μm diameter) micropatterns, the variability is also high but the distribution of the stem cell fraction becomes unimodal. Using a stochastic model, we analyze the differentiation dynamics and quantitatively determine the differentiation probability as a function of stem cell fraction. Results indicate that stem cells can interact and sense cellular composition in their immediate neighborhood and adjust their differentiation probability accordingly. Blocking epithelial cadherin (E-cadherin) can diminish this cell-cell contact mediated sensing. For larger micropatterns, cell motility adds a spatial dimension to the picture. Taken together, we find stochasticity and cell-cell interactions are important factors in determining cell fate in mixed cell populations.
Figures
References
-
- Waddington C. H. The strategy of the genes; a discussion of some aspects of theoretical biology. ix-262 (Allen & Unwin, 1957).
-
- Takahashi K. et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 131, 861–872 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

