Emerging roles of lysine methylation on non-histone proteins
- PMID: 26227335
- PMCID: PMC11114002
- DOI: 10.1007/s00018-015-2001-4
Emerging roles of lysine methylation on non-histone proteins
Abstract
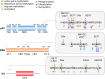
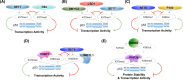
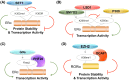
Lysine methylation is a common posttranslational modification (PTM) of histones that is important for the epigenetic regulation of transcription and chromatin in eukaryotes. Increasing evidence demonstrates that in addition to histones, lysine methylation also occurs on various non-histone proteins, especially transcription- and chromatin-regulating proteins. In this review, we will briefly describe the histone lysine methyltransferases (KMTs) that have a broad spectrum of non-histone substrates. We will use p53 and nuclear receptors, especially estrogen receptor alpha, as examples to discuss the dynamic nature of non-histone protein lysine methylation, the writers, erasers, and readers of these modifications, and the crosstalk between lysine methylation and other PTMs in regulating the functions of the modified proteins. Understanding the roles of lysine methylation in normal cells and during development will shed light on the complex biology of diseases associated with the dysregulation of lysine methylation on both histones and non-histone proteins.
Keywords: ERα; G9a; Lysine methylation; SETD7; SMYD2; p53.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

