The frequent evolutionary birth and death of functional promoters in mouse and human
- PMID: 26228054
- PMCID: PMC4579340
- DOI: 10.1101/gr.190546.115
The frequent evolutionary birth and death of functional promoters in mouse and human
Abstract
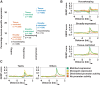
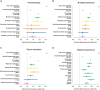
Promoters are central to the regulation of gene expression. Changes in gene regulation are thought to underlie much of the adaptive diversification between species and phenotypic variation within populations. In contrast to earlier work emphasizing the importance of enhancer evolution and subtle sequence changes at promoters, we show that dramatic changes such as the complete gain and loss (collectively, turnover) of functional promoters are common. Using quantitative measures of transcription initiation in both humans and mice across 52 matched tissues, we discriminate promoter sequence gains from losses and resolve the lineage of changes. We also identify expression divergence and functional turnover between orthologous promoters, finding only the latter is associated with local sequence changes. Promoter turnover has occurred at the majority (>56%) of protein-coding genes since humans and mice diverged. Tissue-restricted promoters are the most evolutionarily volatile where retrotransposition is an important, but not the sole, source of innovation. There is considerable heterogeneity of turnover rates between promoters in different tissues, but the consistency of these in both lineages suggests that the same biological systems are similarly inclined to transcriptional rewiring. The genes affected by promoter turnover show evidence of adaptive evolution. In mice, promoters are primarily lost through deletion of the promoter containing sequence, whereas in humans, many promoters appear to be gradually decaying with weak transcriptional output and relaxed selective constraint. Our results suggest that promoter gain and loss is an important process in the evolutionary rewiring of gene regulation and may be a significant source of phenotypic diversification.
© 2015 Young et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Anderson E, Hill RE. 2014. Long range regulation of the sonic hedgehog gene. Curr Opin Genet Dev 27C: 54–59. - PubMed
-
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441: 87–90. - PubMed
-
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
