Why do we study animal toxins?
- PMID: 26228472
- PMCID: PMC4790257
- DOI: 10.13918/j.issn.2095-8137.2015.4.183
Why do we study animal toxins?
Abstract
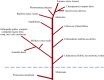
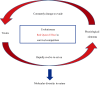
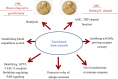
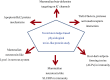
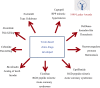
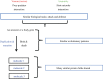
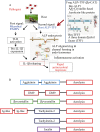
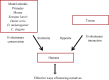
Venom (toxins) is an important trait evolved along the evolutionary tree of animals. Our knowledges on venoms, such as their origins and loss, the biological relevance and the coevolutionary patterns with other organisms are greatly helpful in understanding many fundamental biological questions, i.e., the environmental adaptation and survival competition, the evolution shaped development and balance of venoms, and the sophisticated correlations among venom, immunity, body power, intelligence, their genetic basis, inherent association, as well as the cost-benefit and trade-offs of biological economy. Lethal animal envenomation can be found worldwide. However, from foe to friend, toxin studies have led lots of important discoveries and exciting avenues in deciphering and fighting human diseases, including the works awarded the Nobel Prize and lots of key clinic therapeutics. According to our survey, so far, only less than 0.1% of the toxins of the venomous animals in China have been explored. We emphasize on the similarities shared by venom and immune systems, as well as the studies of toxin knowledge-based physiological toxin-like proteins/peptides (TLPs). We propose the natural pairing hypothesis. Evolution links toxins with humans. Our mission is to find out the right natural pairings and interactions of our body elements with toxins, and with endogenous toxin-like molecules. Although, in nature, toxins may endanger human lives, but from a philosophical point of view, knowing them well is an effective way to better understand ourselves. So, this is why we study toxins.
Keywords: Disease mechanism; Drug development; Evolution; Survival competition; Toxins.
Figures

References
-
- Aili SR, Touchard A, Escoubas P, Padula MP, Orivel J, Dejean A, Nicholson GM.2014. Diversity of peptide toxins from stinging ant venoms.Toxicon, 92: 183-178. - PubMed
-
- Al-Hassan JM, Thomson M, Criddle KR, Summers B, Criddle RS.1985. Catfish epidermal secretions in response to threat or injury.Marine Biology, 88(2): 117-123.
-
- Almaaytah A, Albalas Q.2014. Scorpion venom peptides with no disulfide bridges: a review.Peptides, 51: 35-45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
