Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1
- PMID: 26229009
- PMCID: PMC4572411
- DOI: 10.1152/ajpgi.00140.2015
Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1
Abstract
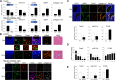
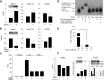
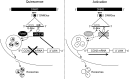
A hallmark of liver fibrosis is the activation of hepatic stellate cells (HSC), which results in their production of fibrotic molecules, a process that is largely regulated by connective tissue growth factor (CCN2). CCN2 is increasingly expressed during HSC activation because of diminished expression of microRNA-214 (miR-214), a product of dynamin 3 opposite strand (DNM3os) that directly suppresses CCN2 mRNA. We show that an E-box in the miR-214 promoter binds the basic helix-loop-helix transcription factor, Twist1, which drives miR-214 expression and results in CCN2 suppression. Twist1 expression was suppressed in HSC of fibrotic livers or in cultured HSC undergoing activation in vitro or after treatment with ethanol. Furthermore, Twist1 decreasingly interacted with DNM3os as HSC underwent activation in vitro. Nanovesicular exosomes secreted by quiescent but not activated HSC contained high levels of Twist1, thus reflecting the suppression of cellular Twist1 during HSC activation. Exosomal Twist1 was intercellularly shuttled between HSC and stimulated expression of miR-214 in the recipient cells, causing expression of CCN2 and its downstream effectors to be suppressed. Additionally, the miR-214 E-box in HSC was also regulated by hepatocyte-derived exosomes, showing that functional transfer of exosomal Twist1 occurs between different cell types. Finally, the levels of Twist1, miR-214, or CCN2 in circulating exosomes from fibrotic mice reflected fibrosis-induced changes in the liver itself, highlighting the potential utility of these and other constituents in serum exosomes as novel circulating biomarkers for liver fibrosis. These findings reveal a unique function for cellular or exosomal Twist1 in CCN2-dependent fibrogenesis.
Keywords: CCN2; E-box; connective tissue growth factor; exosome; fibrosis.
Copyright © 2015 the American Physiological Society.
Figures

References
-
- Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, Perrin-Schmitt F. The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Hum Mol Genet 7: 945–957, 1998. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

