Viruses transfer the antiviral second messenger cGAMP between cells
- PMID: 26229117
- PMCID: PMC4617605
- DOI: 10.1126/science.aab3632
Viruses transfer the antiviral second messenger cGAMP between cells
Abstract
Cyclic GMP-AMP synthase (cGAS) detects cytosolic DNA during virus infection and induces an antiviral state. cGAS signals by synthesis of a second messenger, cyclic GMP-AMP (cGAMP), which activates stimulator of interferon genes (STING). We show that cGAMP is incorporated into viral particles, including lentivirus and herpesvirus virions, when these are produced in cGAS-expressing cells. Virions transferred cGAMP to newly infected cells and triggered a STING-dependent antiviral program. These effects were independent of exosomes and viral nucleic acids. Our results reveal a way by which a signal for innate immunity is transferred between cells, potentially accelerating and broadening antiviral responses. Moreover, infection of dendritic cells with cGAMP-loaded lentiviruses enhanced their activation. Loading viral vectors with cGAMP therefore holds promise for vaccine development.
Copyright © 2015, American Association for the Advancement of Science.
Figures
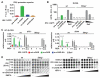


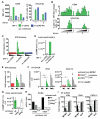
Comment in
-
Inside Job: Viruses Transfer cGAMP between Cells.Cell Host Microbe. 2015 Sep 9;18(3):263-5. doi: 10.1016/j.chom.2015.08.008. Cell Host Microbe. 2015. PMID: 26355209
-
IMMUNOLOGY. Viruses carry antiviral cargo.Science. 2015 Sep 11;349(6253):1166-7. doi: 10.1126/science.aad0942. Science. 2015. PMID: 26359389 No abstract available.
References
-
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

