Differential Proteomic Analysis Using iTRAQ Reveals Alterations in Hull Development in Rice (Oryza sativa L.)
- PMID: 26230730
- PMCID: PMC4521873
- DOI: 10.1371/journal.pone.0133696
Differential Proteomic Analysis Using iTRAQ Reveals Alterations in Hull Development in Rice (Oryza sativa L.)
Abstract
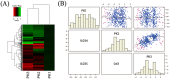
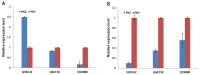
Rice hull, the outer cover of the rice grain, determines grain shape and size. Changes in the rice hull proteome in different growth stages may reflect the underlying mechanisms involved in grain development. To better understand these changes, isobaric tags for relative and absolute quantitative (iTRAQ) MS/MS was used to detect statistically significant changes in the rice hull proteome in the booting, flowering, and milk-ripe growth stages. Differentially expressed proteins were analyzed to predict their potential functions during development. Gene ontology (GO) terms and pathways were used to evaluate the biological mechanisms involved in rice hull at the three growth stages. In total, 5,268 proteins were detected and characterized, of which 563 were differentially expressed across the development stages. The results showed that the flowering and milk-ripe stage proteomes were more similar to each other (r=0.61) than either was to the booting stage proteome. A GO enrichment analysis of the differentially expressed proteins was used to predict their roles during rice hull development. The potential functions of 25 significantly differentially expressed proteins were used to evaluate their possible roles at various growth stages. Among these proteins, an unannotated protein (Q7X8A1) was found to be overexpressed especially in the flowering stage, while a putative uncharacterized protein (B8BF94) and an aldehyde dehydrogenase (Q9FPK6) were overexpressed only in the milk-ripe stage. Pathways regulated by differentially expressed proteins were also analyzed. Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase (Q9SDJ2), and two magnesium-chelatase subunits, ChlD (Q6ATS0), and ChlI (Q53RM0), were associated with chlorophyll biosynthesis at different developmental stages. The expression of Q9SDJ2 in the flowering and milk-ripe stages was validated by qRT-PCR. The 25 candidate proteins may be pivotal markers for controlling rice hull development at various growth stages and chlorophyll biosynthesis pathway related proteins, especially magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase (Q9SDJ2), may provide new insights into the molecular mechanisms of rice hull development and chlorophyll associated regulation.
Conflict of interest statement
Figures



Similar articles
-
Comparative proteomic analysis reveals alterations in development and photosynthesis-related proteins in diploid and triploid rice.BMC Plant Biol. 2016 Sep 13;16(1):199. doi: 10.1186/s12870-016-0891-4. BMC Plant Biol. 2016. PMID: 27619227 Free PMC article.
-
iTRAQ-Based Quantitative Proteomics Analysis of Black Rice Grain Development Reveals Metabolic Pathways Associated with Anthocyanin Biosynthesis.PLoS One. 2016 Jul 14;11(7):e0159238. doi: 10.1371/journal.pone.0159238. eCollection 2016. PLoS One. 2016. PMID: 27415428 Free PMC article.
-
Proteomic Analysis Reveals Coordinated Regulation of Anthocyanin Biosynthesis through Signal Transduction and Sugar Metabolism in Black Rice Leaf.Int J Mol Sci. 2017 Dec 15;18(12):2722. doi: 10.3390/ijms18122722. Int J Mol Sci. 2017. PMID: 29244752 Free PMC article.
-
Rice proteomics: current status and future perspectives.Electrophoresis. 2003 Oct;24(19-20):3378-89. doi: 10.1002/elps.200305586. Electrophoresis. 2003. PMID: 14595685 Review.
-
Molecular functions of genes related to grain shape in rice.Breed Sci. 2015 Mar;65(2):120-6. doi: 10.1270/jsbbs.65.120. Epub 2015 Mar 1. Breed Sci. 2015. PMID: 26069441 Free PMC article. Review.
Cited by
-
Comparative proteomic analysis reveals alterations in development and photosynthesis-related proteins in diploid and triploid rice.BMC Plant Biol. 2016 Sep 13;16(1):199. doi: 10.1186/s12870-016-0891-4. BMC Plant Biol. 2016. PMID: 27619227 Free PMC article.
-
PSII Activity Was Inhibited at Flowering Stage with Developing Black Bracts of Oat.Int J Mol Sci. 2021 May 17;22(10):5258. doi: 10.3390/ijms22105258. Int J Mol Sci. 2021. PMID: 34067635 Free PMC article.
-
Global Proteomic Analysis Reveals Widespread Lysine Succinylation in Rice Seedlings.Int J Mol Sci. 2019 Nov 25;20(23):5911. doi: 10.3390/ijms20235911. Int J Mol Sci. 2019. PMID: 31775301 Free PMC article.
-
iTRAQ-Based Proteomics Analysis and Network Integration for Kernel Tissue Development in Maize.Int J Mol Sci. 2017 Aug 24;18(9):1840. doi: 10.3390/ijms18091840. Int J Mol Sci. 2017. PMID: 28837076 Free PMC article.
References
-
- Guo S-b, Wei Y, Li X-q, Liu K-q, Huang F-k, Chen C-h, et al. (2013) Development and Identification of Introgression Lines from Cross of Oryza sativa and Oryza minuta. Rice Science 20: 95–102.
-
- Lu C, Liu J, Jiang J-h, BRERIA CM, Tan H-l, ICHII M, et al. (2013) Development and Application of SCAR Markers for Discriminating Cytoplasmic Male Sterile Lines from Their Cognate Maintainer Lines in Indica Rice. Rice Science 20: 191–199.
-
- Taiz L and Zeiger E (2012) Plant Physiology, Third Edition Sinauer Associates, Sunderland, MA: 484.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

