An international data set for CMML validates prognostic scoring systems and demonstrates a need for novel prognostication strategies
- PMID: 26230957
- PMCID: PMC4526779
- DOI: 10.1038/bcj.2015.53
An international data set for CMML validates prognostic scoring systems and demonstrates a need for novel prognostication strategies
Abstract
Since its reclassification as a distinct disease entity, clinical research efforts have attempted to establish baseline characteristics and prognostic scoring systems for chronic myelomonocytic leukemia (CMML). Although existing data for baseline characteristics and CMML prognostication have been robustly developed and externally validated, these results have been limited by the small size of single-institution cohorts. We developed an international CMML data set that included 1832 cases across eight centers to establish the frequency of key clinical characteristics. Of note, we found that the majority of CMML patients were classified as World Health Organization CMML-1 and that a 7.5% bone marrow blast cut-point may discriminate prognosis with higher resolution in comparison with the existing 10%. We additionally interrogated existing CMML prognostic models and found that they are all valid and have comparable performance but are vulnerable to upstaging. Using random forest survival analysis for variable discovery, we demonstrated that the prognostic power of clinical variables alone is limited. Last, we confirmed the independent prognostic relevance of ASXL1 gene mutations and identified the novel adverse prognostic impact imparted by CBL mutations. Our data suggest that combinations of clinical and molecular information may be required to improve the accuracy of current CMML prognostication.
Figures
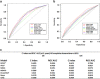
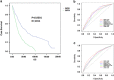
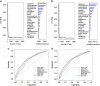

References
-
- Padron E, Komrokji R, List AF. The clinical management of chronic myelomonocytic leukemia. Clin Adv Hematol Oncol. 2014;12:172–178. - PubMed
-
- Bennett JM, Catovsky D, Daniel M-T, Flandrin G, Galton DAG, Gralnick HR, et al. Proposals for the classification of the acute leukaemias French–American–British (FAB) Co-operative Group. Br J Haematol. 1976;33:451–458. - PubMed
-
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

