Loss of heterozygosity: what is it good for?
- PMID: 26231170
- PMCID: PMC4522148
- DOI: 10.1186/s12920-015-0123-z
Loss of heterozygosity: what is it good for?
Abstract
Background: Loss of heterozygosity (LOH) is a common genetic event in cancer development, and is known to be involved in the somatic loss of wild-type alleles in many inherited cancer syndromes. The wider involvement of LOH in cancer is assumed to relate to unmasking a somatically mutated tumour suppressor gene through loss of the wild type allele.
Methods: We analysed 86 ovarian carcinomas for mutations in 980 genes selected on the basis of their location in common regions of LOH.
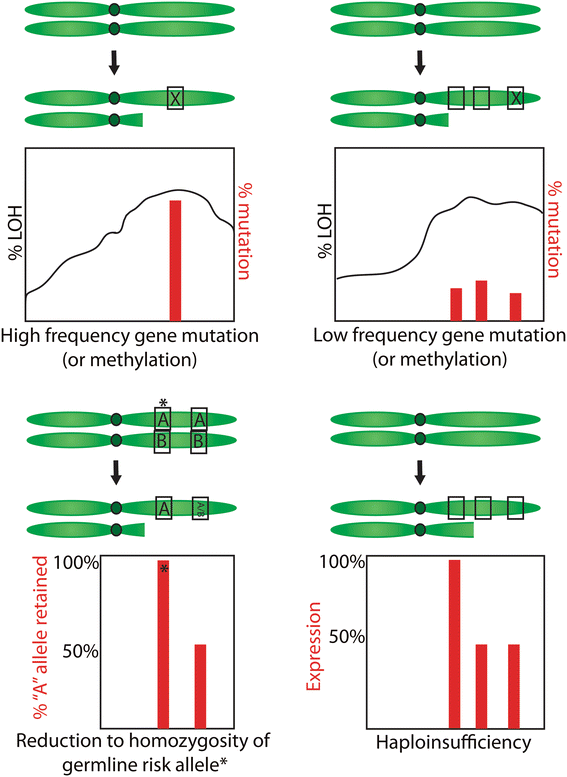
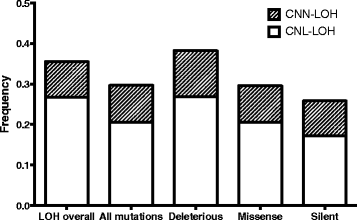
Results: We identified 36 significantly mutated genes, but these could only partly account for the quanta of LOH in the samples. Using our own and TCGA data we then evaluated five possible models to explain the selection for non-random accumulation of LOH in ovarian cancer genomes: 1. Classic two-hit hypothesis: high frequency biallelic genetic inactivation of tumour suppressor genes. 2. Epigenetic two-hit hypothesis: biallelic inactivation through methylation and LOH. 3. Multiple alternate-gene biallelic inactivation: low frequency gene disruption. 4. Haplo-insufficiency: Single copy gene disruption. 5. Modified two-hit hypothesis: reduction to homozygosity of low penetrance germline predisposition alleles. We determined that while high-frequency biallelic gene inactivation under model 1 is rare, regions of LOH (particularly copy-number neutral LOH) are enriched for deleterious mutations and increased promoter methylation, while copy-number loss LOH regions are likely to contain under-expressed genes suggestive of haploinsufficiency. Reduction to homozygosity of cancer predisposition SNPs may also play a minor role.
Conclusion: It is likely that selection for regions of LOH depends on its effect on multiple genes. Selection for copy number neutral LOH may better fit the classic two-hit model whereas selection for copy number loss may be attributed to its effect on multi-gene haploinsufficiency. LOH mapping alone is unlikely to be successful in identifying novel tumour suppressor genes; a combined approach may be more effective.
Figures


References
-
- Merajver SD, Frank TS, Xu J, Pham TM, Calzone KA, Bennett-Baker P, et al. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin Cancer Res. 1995;1(5):539–44. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

