Hansenula polymorpha Hac1p Is Critical to Protein N-Glycosylation Activity Modulation, as Revealed by Functional and Transcriptomic Analyses
- PMID: 26231645
- PMCID: PMC4579441
- DOI: 10.1128/AEM.01440-15
Hansenula polymorpha Hac1p Is Critical to Protein N-Glycosylation Activity Modulation, as Revealed by Functional and Transcriptomic Analyses
Abstract
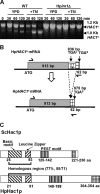
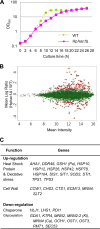
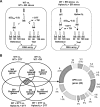
Aggregation of misfolded protein in the endoplasmic reticulum (ER) induces a cellular protective response to ER stress, the unfolded protein response (UPR), which is mediated by a basic leucine zipper (bZIP) transcription factor, Hac1p/Xbp1. In this study, we identified and studied the molecular functions of a HAC1 homolog from the thermotolerant yeast Hansenula polymorpha (HpHAC1). We found that the HpHAC1 mRNA contains a nonconventional intron of 177 bp whose interaction with the 5' untranslated region is responsible for the translational inhibition of the HpHAC1 mRNA. The H. polymorpha hac1-null (Hphac1Δ) mutant strain grew slowly, even under normal growth conditions, and was less thermotolerant than the wild-type (WT) strain. The mutant strain was also more sensitive to cell wall-perturbing agents and to the UPR-inducing agents dithiothreitol (DTT) and tunicamycin (TM). Using comparative transcriptome analysis of the WT and Hphac1Δ strains treated with DTT and TM, we identified HpHAC1-dependent core UPR targets, which included genes involved in protein secretion and processing, particularly those required for N-linked protein glycosylation. Notably, different glycosylation and processing patterns of the vacuolar glycoprotein carboxypeptidase Y were observed in the WT and Hphac1Δ strains. Moreover, overexpression of active HpHac1p significantly increased the N-linked glycosylation efficiency and TM resistance. Collectively, our results suggest that the function of HpHac1p is important not only for UPR induction but also for efficient glycosylation in H. polymorpha.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

