Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages
- PMID: 26232173
- PMCID: PMC4566811
- DOI: 10.1182/blood-2015-01-624809
Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages
Abstract
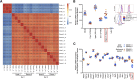
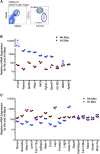
Alveolar macrophages (AMs) reside on the luminal surfaces of the airways and alveoli where they maintain host defense and promote alveolar homeostasis by ingesting inhaled particulates and regulating inflammatory responses. Recent studies have demonstrated that AMs populate the lungs during embryogenesis and self-renew throughout life with minimal replacement by circulating monocytes, except under extreme conditions of depletion or radiation injury. Here we demonstrate that on a global scale, environment appears to dictate AM development and function. Indeed, transcriptome analysis of embryonic host-derived and postnatal donor-derived AMs coexisting within the same mouse demonstrated >98% correlation and overall functional analyses were similar. However, we also identified several genes whose expression was dictated by origin rather than environment. The most differentially expressed gene not altered by environment was Marco, a gene recently demonstrated to have enhancer activity in embryonic-derived but not postnatal-derived tissue macrophages. Overall, we show that under homeostatic conditions, the environment largely dictates the programming and function of AMs, whereas the expression of a small number of genes remains linked to the origin of the cell.
© 2015 by The American Society of Hematology.
Figures



References
-
- Thepen T, Kraal G, Holt PG. The role of alveolar macrophages in regulation of lung inflammation. Ann N Y Acad Sci. 1994;725:200–206. - PubMed
-
- Guilliams M, Lambrecht BN, Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013;6(3):464–473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

