A method for systematic discovery of adverse drug events from clinical notes
- PMID: 26232442
- PMCID: PMC4921953
- DOI: 10.1093/jamia/ocv102
A method for systematic discovery of adverse drug events from clinical notes
Abstract
Objective: Adverse drug events (ADEs) are undesired harmful effects resulting from use of a medication, and occur in 30% of hospitalized patients. The authors have developed a data-mining method for systematic, automated detection of ADEs from electronic medical records.
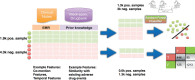
Materials and methods: This method uses the text from 9.5 million clinical notes, along with prior knowledge of drug usages and known ADEs, as inputs. These inputs are further processed into statistics used by a discriminative classifier which outputs the probability that a given drug-disorder pair represents a valid ADE association. Putative ADEs identified by the classifier are further filtered for positive support in 2 independent, complementary data sources. The authors evaluate this method by assessing support for the predictions in other curated data sources, including a manually curated, time-indexed reference standard of label change events.
Results: This method uses a classifier that achieves an area under the curve of 0.94 on a held out test set. The classifier is used on 2,362,950 possible drug-disorder pairs comprised of 1602 unique drugs and 1475 unique disorders for which we had data, resulting in 240 high-confidence, well-supported drug-AE associations. Eighty-seven of them (36%) are supported in at least one of the resources that have information that was not available to the classifier.
Conclusion: This method demonstrates the feasibility of systematic post-marketing surveillance for ADEs using electronic medical records, a key component of the learning healthcare system.
Keywords: EMR mining; machine learning; pharmacovigilance; post market drug safety surveillance.
© The Author 2015. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277(4):301–306. - PubMed
-
- Classen DC, Resar R, Griffin F, et al. ‘Global trigger tool' shows that adverse events in hospitals may be ten times greater than previously measured. Health Aff. 2011;30(4):581–589. - PubMed
-
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279(15):1200–1205. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

