Protein structure refinement via molecular-dynamics simulations: What works and what does not?
- PMID: 26234208
- PMCID: PMC5493977
- DOI: 10.1002/prot.24871
Protein structure refinement via molecular-dynamics simulations: What works and what does not?
Abstract
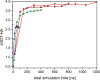
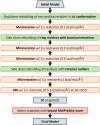
Protein structure refinement during CASP11 by the Feig group was described. Molecular dynamics simulations were used in combination with an improved selection and averaging protocol. On average, modest refinement was achieved with some targets improved significantly. Analysis of the CASP submission from our group focused on refinement success versus amount of sampling, refinement of different secondary structure elements and whether refinement varied as a function of which group provided initial models. The refinement of local stereochemical features was examined via the MolProbity score and an updated protocol was developed that can generate high-quality structures with very low MolProbity scores for most starting structures with modest computational effort. Proteins 2016; 84(Suppl 1):282-292. © 2015 Wiley Periodicals, Inc.
Keywords: CASP; Molprobity; protein; scoring; structure prediction.
© 2015 Wiley Periodicals, Inc.
Figures




References
-
- Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How Fast-Folding Proteins Fold. Science. 2011;334:517–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

