Energy coupling mechanisms of MFS transporters
- PMID: 26234418
- PMCID: PMC4594656
- DOI: 10.1002/pro.2759
Energy coupling mechanisms of MFS transporters
Abstract
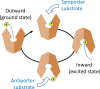
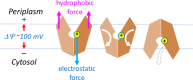
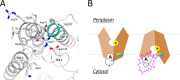
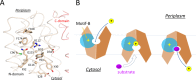
Major facilitator superfamily (MFS) is a large class of secondary active transporters widely expressed across all life kingdoms. Although a common 12-transmembrane helix-bundle architecture is found in most MFS crystal structures available, a common mechanism of energy coupling remains to be elucidated. Here, we discuss several models for energy-coupling in the transport process of the transporters, largely based on currently available structures and the results of their biochemical analyses. Special attention is paid to the interaction between protonation and the negative-inside membrane potential. Also, functional roles of the conserved sequence motifs are discussed in the context of the 3D structures. We anticipate that in the near future, a unified picture of the functions of MFS transporters will emerge from the insights gained from studies of the common architectures and conserved motifs.
Keywords: MFS transporters; energy coupling mechanisms; membrane potential; motif A.
© 2015 The Protein Society.
Figures



References
-
- Marger MD, Saier MH., Jr. ( A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. - PubMed
-
- Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS( The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. - PubMed
-
- Poolman B, Konings WN( Secondary solute transport in bacteria. Biochim Biophys Acta. 1993;1183:5–39. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

