Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells
- PMID: 26234680
- PMCID: PMC4650269
- DOI: 10.1038/onc.2015.270
Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells
Abstract
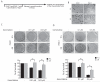
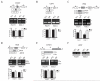
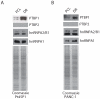
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive and incurable disease. Poor prognosis is due to multiple reasons, including acquisition of resistance to gemcitabine, the first-line chemotherapeutic approach. Thus, there is a strong need for novel therapies, targeting more directly the molecular aberrations of this disease. We found that chronic exposure of PDAC cells to gemcitabine selected a subpopulation of cells that are drug-resistant (DR-PDAC cells). Importantly, alternative splicing (AS) of the pyruvate kinase gene (PKM) was differentially modulated in DR-PDAC cells, resulting in promotion of the cancer-related PKM2 isoform, whose high expression also correlated with shorter recurrence-free survival in PDAC patients. Switching PKM splicing by antisense oligonucleotides to favor the alternative PKM1 variant rescued sensitivity of DR-PDAC cells to gemcitabine and cisplatin, suggesting that PKM2 expression is required to withstand drug-induced genotoxic stress. Mechanistically, upregulation of the polypyrimidine-tract binding protein (PTBP1), a key modulator of PKM splicing, correlated with PKM2 expression in DR-PDAC cell lines. PTBP1 was recruited more efficiently to PKM pre-mRNA in DR- than in parental PDAC cells. Accordingly, knockdown of PTBP1 in DR-PDAC cells reduced its recruitment to the PKM pre-mRNA, promoted splicing of the PKM1 variant and abolished drug resistance. Thus, chronic exposure to gemcitabine leads to upregulation of PTBP1 and modulation of PKM AS in PDAC cells, conferring resistance to the drug. These findings point to PKM2 and PTBP1 as new potential therapeutic targets to improve response of PDAC to chemotherapy.
Figures


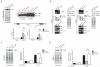
References
-
- Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7(3):163–72. - PubMed
-
- Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62(2):317–26. - PubMed
-
- Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

