STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease
- PMID: 26235147
- PMCID: PMC4537353
- DOI: 10.1016/j.chom.2015.07.001
STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease
Abstract
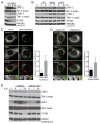
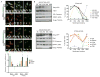
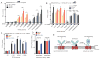
STING is an ER-associated membrane protein that is critical for innate immune sensing of pathogens. STING-mediated activation of the IFN-I pathway through the TBK1/IRF3 signaling axis involves both cyclic-dinucleotide binding and its translocation from the ER to vesicles. However, how these events are coordinated, and the exact mechanism of STING activation, remain poorly understood. Here, we found that the Shigella effector protein IpaJ potently inhibits STING signaling by blocking its translocation from the ER to ERGIC, even in the context of dinucleotide binding. Reconstitution using purified components revealed STING translocation as the rate-limiting event in maximal signal transduction. Furthermore, STING mutations associated with autoimmunity in humans were found to cause constitutive ER exit and to activate STING independent of cGAMP binding. Together, these data provide compelling evidence for an ER retention and ERGIC/Golgi-trafficking mechanism of STING regulation that is subverted by bacterial pathogens and is deregulated in human genetic disease.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest.
Figures



Comment in
-
STING Signaling the enERGIC Way.Cell Host Microbe. 2015 Aug 12;18(2):137-9. doi: 10.1016/j.chom.2015.07.014. Cell Host Microbe. 2015. PMID: 26269948
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

