Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer
- PMID: 26235388
- PMCID: PMC4536608
- DOI: 10.1186/s13059-015-0719-9
Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer
Abstract
Background: Epigenetic changes are being increasingly recognized as a prominent feature of cancer. This occurs not only at individual genes, but also over larger chromosomal domains. To investigate this, we set out to identify large chromosomal domains of epigenetic dysregulation in breast cancers.
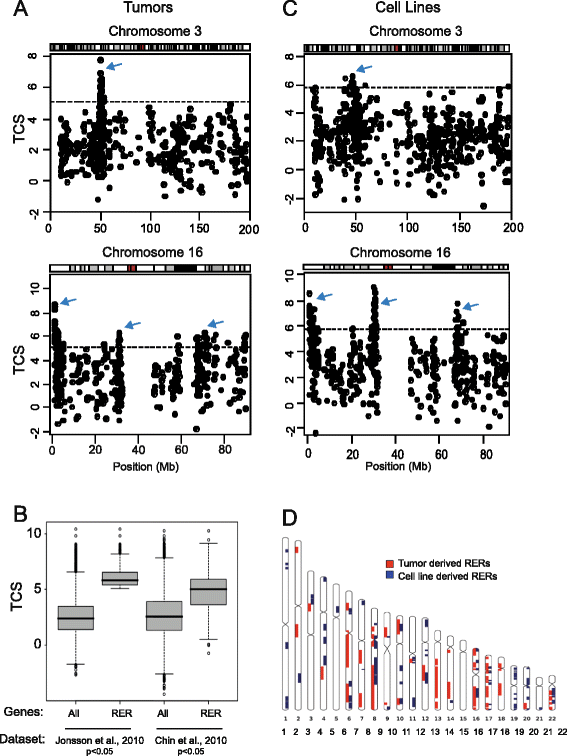
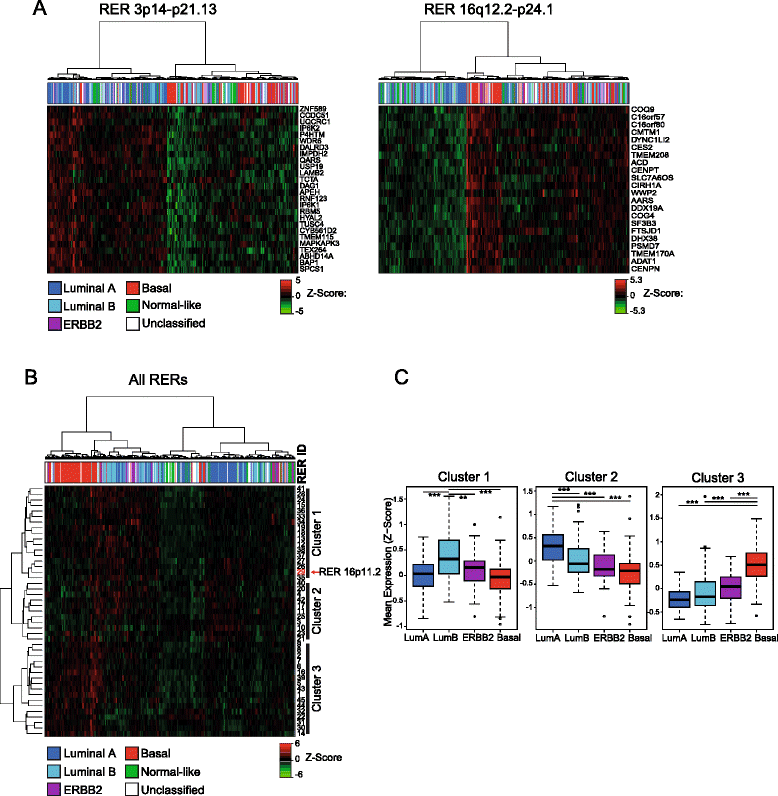
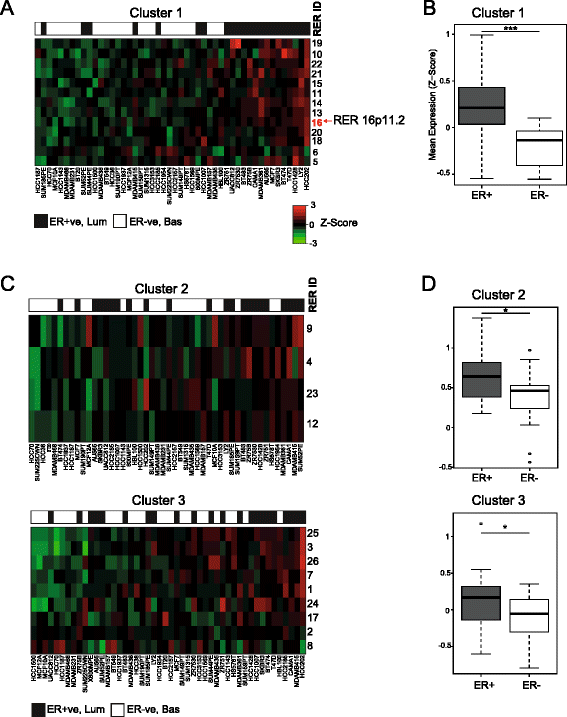
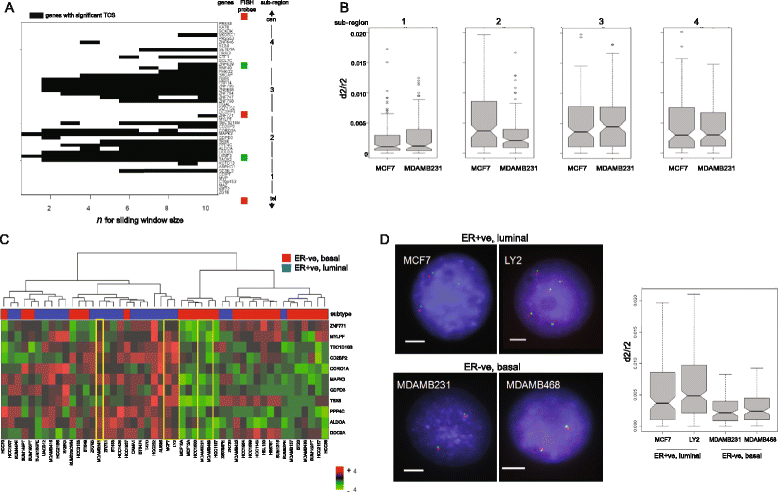
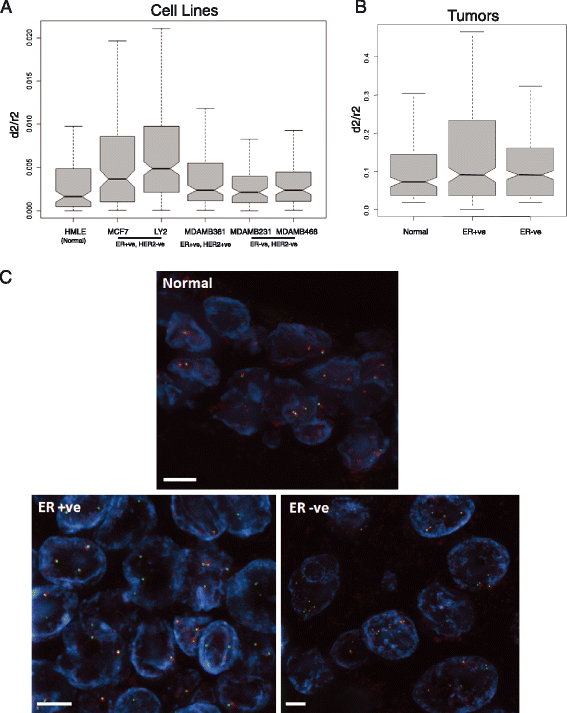
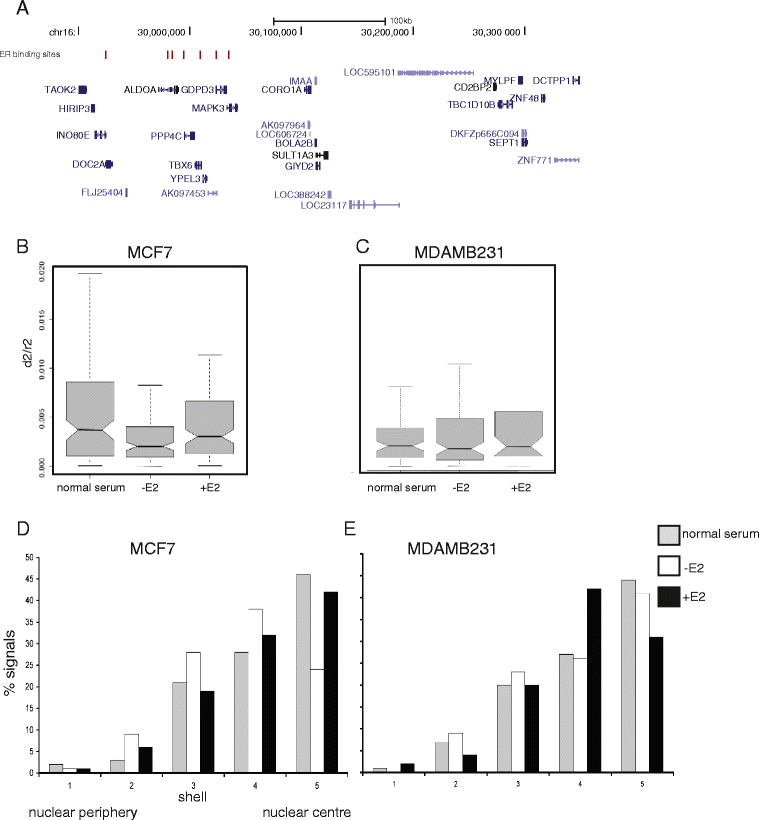
Results: We identify large regions of coordinate down-regulation of gene expression, and other regions of coordinate activation, in breast cancers and show that these regions are linked to tumor subtype. In particular we show that a group of coordinately regulated regions are expressed in luminal, estrogen-receptor positive breast tumors and cell lines. For one of these regions of coordinate gene activation, we show that regional epigenetic regulation is accompanied by visible unfolding of large-scale chromatin structure and a repositioning of the region within the nucleus. In MCF7 cells, we show that this depends on the presence of estrogen.
Conclusions: Our data suggest that the liganded estrogen receptor is linked to long-range changes in higher-order chromatin organization and epigenetic dysregulation in cancer. This may suggest that as well as drugs targeting histone modifications, it will be valuable to investigate the inhibition of protein complexes involved in chromatin folding in cancer cells.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

