Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria
- PMID: 26235513
- PMCID: PMC4532887
- DOI: 10.1038/ncomms8922
Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria
Abstract
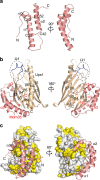
Eukaryotic cells are compartmentalized into membrane-bounded organelles whose functions rely on lipid trafficking to achieve membrane-specific compositions of lipids. Here we focused on the Ups1-Mdm35 system, which mediates phosphatidic acid (PA) transfer between the outer and inner mitochondrial membranes, and determined the X-ray structures of Mdm35 and Ups1-Mdm35 with and without PA. The Ups1-Mdm35 complex constitutes a single domain that has a deep pocket and flexible Ω-loop lid. Structure-based mutational analyses revealed that a basic residue at the pocket bottom and the Ω-loop lid are important for PA extraction from the membrane following Ups1 binding. Ups1 binding to the membrane is enhanced by the dissociation of Mdm35. We also show that basic residues around the pocket entrance are important for Ups1 binding to the membrane and PA extraction. These results provide a structural basis for understanding the mechanism of PA transfer between mitochondrial membranes.
Figures




Similar articles
-
Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex.EMBO Rep. 2015 Jul;16(7):813-23. doi: 10.15252/embr.201540137. Epub 2015 Jun 12. EMBO Rep. 2015. PMID: 26071601 Free PMC article.
-
Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.Commun Biol. 2020 Aug 25;3(1):468. doi: 10.1038/s42003-020-01121-x. Commun Biol. 2020. PMID: 32843686 Free PMC article.
-
Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein.Science. 2012 Nov 9;338(6108):815-8. doi: 10.1126/science.1225625. Epub 2012 Oct 4. Science. 2012. PMID: 23042293
-
Structural comparison of yeast and human intra-mitochondrial lipid transport systems.Biochem Soc Trans. 2016 Apr 15;44(2):479-85. doi: 10.1042/BST20150264. Biochem Soc Trans. 2016. PMID: 27068958 Review.
-
Mitochondrial lipid trafficking.Trends Cell Biol. 2014 Jan;24(1):44-52. doi: 10.1016/j.tcb.2013.07.011. Epub 2013 Sep 1. Trends Cell Biol. 2014. PMID: 24001776 Review.
Cited by
-
Chicken or Egg? Mitochondrial Phospholipids and Oxidative Stress in Disuse-Induced Skeletal Muscle Atrophy.Antioxid Redox Signal. 2023 Feb;38(4-6):338-351. doi: 10.1089/ars.2022.0151. Antioxid Redox Signal. 2023. PMID: 36301935 Free PMC article. Review.
-
The conformational plasticity of structurally unrelated lipid transport proteins correlates with their mode of action.PLoS Biol. 2024 Aug 19;22(8):e3002737. doi: 10.1371/journal.pbio.3002737. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39159271 Free PMC article.
-
The UbiB family member Cqd1 forms a novel membrane contact site in mitochondria.J Cell Sci. 2023 May 15;136(10):jcs260578. doi: 10.1242/jcs.260578. Epub 2023 May 18. J Cell Sci. 2023. PMID: 37073556 Free PMC article.
-
Phospholipid ebb and flow makes mitochondria go.J Cell Biol. 2020 Aug 3;219(8):e202003131. doi: 10.1083/jcb.202003131. J Cell Biol. 2020. PMID: 32614384 Free PMC article. Review.
-
Transport Pathways That Contribute to the Cellular Distribution of Phosphatidylserine.Front Cell Dev Biol. 2021 Sep 1;9:737907. doi: 10.3389/fcell.2021.737907. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34540851 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

