In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector
- PMID: 26235624
- PMCID: PMC4536165
- DOI: 10.1016/j.celrep.2015.07.019
In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector
Abstract
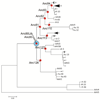
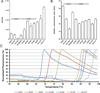
Adeno-associated virus (AAV) vectors have emerged as a gene-delivery platform with demonstrated safety and efficacy in a handful of clinical trials for monogenic disorders. However, limitations of the current generation vectors often prevent broader application of AAV gene therapy. Efforts to engineer AAV vectors have been hampered by a limited understanding of the structure-function relationship of the complex multimeric icosahedral architecture of the particle. To develop additional reagents pertinent to further our insight into AAVs, we inferred evolutionary intermediates of the viral capsid using ancestral sequence reconstruction. In-silico-derived sequences were synthesized de novo and characterized for biological properties relevant to clinical applications. This effort led to the generation of nine functional putative ancestral AAVs and the identification of Anc80, the predicted ancestor of the widely studied AAV serotypes 1, 2, 8, and 9, as a highly potent in vivo gene therapy vector for targeting liver, muscle, and retina.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
LHV and EZ are inventors on a patent describing some of the methods and reagents described here. LHV is inventor on several patents on gene therapy technologies. LHV is co-founder, shareholder, member of the Scientific Advisory Board, and consultant of GenSight Biologics, an ophthalmology gene therapy company.
Figures




References
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Systematic biology. 2006;55:539–552. - PubMed
-
- Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire AM, Wilson JM, Bennett J. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum Mol Genet. 2001;10:3075–3081. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

