The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors
- PMID: 26235645
- PMCID: PMC4522658
- DOI: 10.1038/srep12836
The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors
Abstract
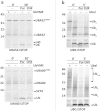
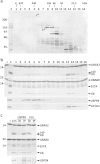
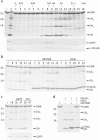
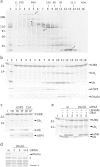
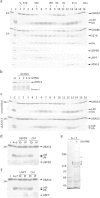
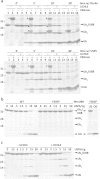
Protein ubiquitination, a major post-translational modification in eukaryotes, requires an adequate pool of free ubiquitin. Cells maintain this pool by two pathways, both involving deubiquitinases (DUBs): recycling of ubiquitin from ubiquitin conjugates and processing of ubiquitin precursors synthesized de novo. Although many advances have been made in recent years regarding ubiquitin recycling, our knowledge on ubiquitin precursor processing is still limited, and questions such as when are these precursors processed and which DUBs are involved remain largely unanswered. Here we provide data suggesting that two of the four mammalian ubiquitin precursors, UBA52 and UBA80, are processed mostly post-translationally whereas the other two, UBB and UBC, probably undergo a combination of co- and post-translational processing. Using an unbiased biochemical approach we found that UCHL3, USP9X, USP7, USP5 and Otulin/Gumby/FAM105b are by far the most active DUBs acting on these precursors. The identification of these DUBs together with their properties suggests that each ubiquitin precursor can be processed in at least two different manners, explaining the robustness of the ubiquitin de novo synthesis pathway.
Figures

References
-
- Amm I., Sommer T. & Wolf D. H. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196 (2014). - PubMed
-
- van Cuijk L., Vermeulen W. & Marteijn J. A. Ubiquitin at work: The ubiquitous regulation of the damage recognition step of NER. Exp. Cell Res. 329, 101–109(2014). - PubMed
-
- Francisco T. et al. Ubiquitin in the peroxisomal protein import pathway. Biochimie 98, 29–35 (2014). - PubMed
-
- Tanno H. & Komada M. The ubiquitin code and its decoding machinery in the endocytic pathway. J. Biochem. 153, 497–504 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

