Mutations in TBX18 Cause Dominant Urinary Tract Malformations via Transcriptional Dysregulation of Ureter Development
- PMID: 26235987
- PMCID: PMC4862256
- DOI: 10.1016/j.ajhg.2015.07.001
Mutations in TBX18 Cause Dominant Urinary Tract Malformations via Transcriptional Dysregulation of Ureter Development
Abstract
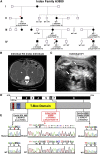
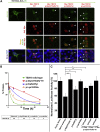
Congenital anomalies of the kidneys and urinary tract (CAKUT) are the most common cause of chronic kidney disease in the first three decades of life. Identification of single-gene mutations that cause CAKUT permits the first insights into related disease mechanisms. However, for most cases the underlying defect remains elusive. We identified a kindred with an autosomal-dominant form of CAKUT with predominant ureteropelvic junction obstruction. By whole exome sequencing, we identified a heterozygous truncating mutation (c.1010delG) of T-Box transcription factor 18 (TBX18) in seven affected members of the large kindred. A screen of additional families with CAKUT identified three families harboring two heterozygous TBX18 mutations (c.1570C>T and c.487A>G). TBX18 is essential for developmental specification of the ureteric mesenchyme and ureteric smooth muscle cells. We found that all three TBX18 altered proteins still dimerized with the wild-type protein but had prolonged protein half life and exhibited reduced transcriptional repression activity compared to wild-type TBX18. The p.Lys163Glu substitution altered an amino acid residue critical for TBX18-DNA interaction, resulting in impaired TBX18-DNA binding. These data indicate that dominant-negative TBX18 mutations cause human CAKUT by interference with TBX18 transcriptional repression, thus implicating ureter smooth muscle cell development in the pathogenesis of human CAKUT.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Smith J.M., Stablein D.M., Munoz R., Hebert D., McDonald R.A. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr. Transplant. 2007;11:366–373. - PubMed
-
- Woolf A.S. A molecular and genetic view of human renal and urinary tract malformations. Kidney Int. 2000;58:500–512. - PubMed
-
- Pohl M., Bhatnagar V., Mendoza S.A., Nigam S.K. Toward an etiological classification of developmental disorders of the kidney and upper urinary tract. Kidney Int. 2002;61:10–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK096238/DK/NIDDK NIH HHS/United States
- 5U54HG006504/HG/NHGRI NIH HHS/United States
- R01 DK088767/DK/NIDDK NIH HHS/United States
- 066647/WT_/Wellcome Trust/United Kingdom
- P30 DK079310/DK/NIDDK NIH HHS/United States
- R01 DK046718/DK/NIDDK NIH HHS/United States
- DK088767/DK/NIDDK NIH HHS/United States
- R01 DK096238/DK/NIDDK NIH HHS/United States
- R56 DK046718/DK/NIDDK NIH HHS/United States
- DK078226/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- DH_/Department of Health/United Kingdom
- R01 DK078226/DK/NIDDK NIH HHS/United States
- R56 DK096238/DK/NIDDK NIH HHS/United States
- U54 HG006504/HG/NHGRI NIH HHS/United States
- MR/L002744/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G0600040/MRC_/Medical Research Council/United Kingdom
- DK46718/DK/NIDDK NIH HHS/United States
- R01 DK103184/DK/NIDDK NIH HHS/United States
- R37 DK046718/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

