The BisPCR(2) method for targeted bisulfite sequencing
- PMID: 26236400
- PMCID: PMC4522100
- DOI: 10.1186/s13072-015-0020-x
The BisPCR(2) method for targeted bisulfite sequencing
Abstract
Background: DNA methylation has emerged as an important regulator of development and disease, necessitating the design of more efficient and cost-effective methods for detecting and quantifying this epigenetic modification. Next-generation sequencing (NGS) techniques offer single base resolution of CpG methylation levels with high statistical significance, but are also high cost if performed genome-wide. Here, we describe a simplified targeted bisulfite sequencing approach in which DNA sequencing libraries are prepared following sodium bisulfite conversion and two rounds of PCR for target enrichment and sample barcoding, termed BisPCR(2).
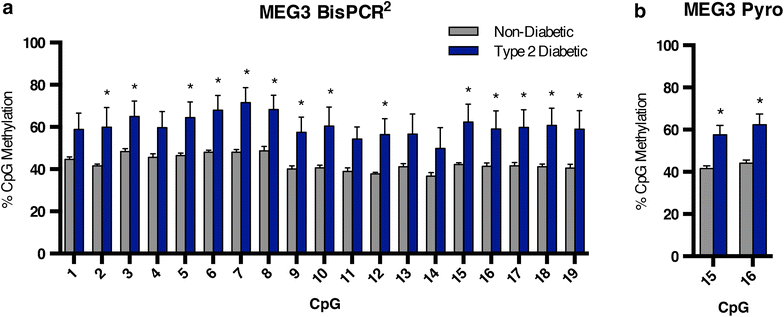
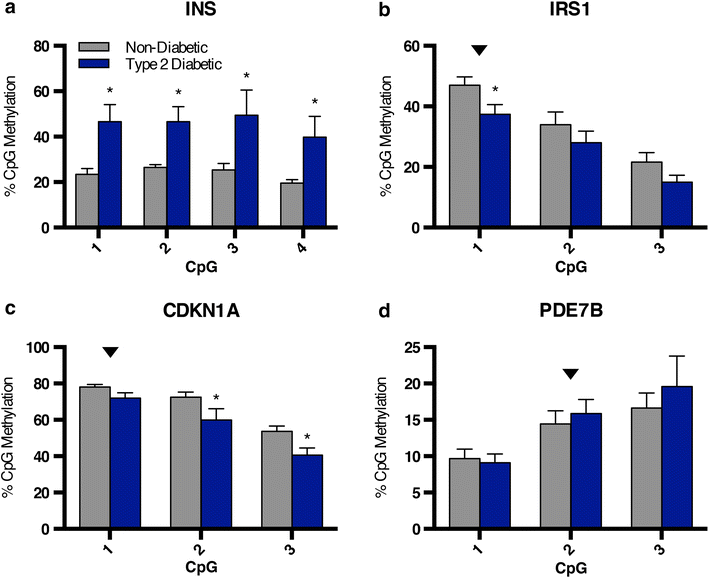
Results: We have applied the BisPCR(2) technique to validate differential methylation at several type 2 diabetes risk loci identified in genome-wide studies of human islets. We confirmed some previous findings while not others, in addition to identifying novel differentially methylated CpGs at these genes of interest, due to the much higher depth of sequencing coverage in BisPCR(2) compared to prior array-based approaches.
Conclusion: This study presents a robust, efficient, and cost-effective technique for targeted bisulfite NGS, and illustrates its utility by reanalysis of prior findings from genome-wide studies.
Keywords: DNA methylation; Next-generation sequencing; Targeted bisulfite sequencing.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

