Comparative genomics of pectinacetylesterases: Insight on function and biology
- PMID: 26237162
- PMCID: PMC4883895
- DOI: 10.1080/15592324.2015.1055434
Comparative genomics of pectinacetylesterases: Insight on function and biology
Erratum in
- doi: 10.1007/s00425-014-2139-6
Abstract
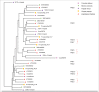
Pectin acetylation influences the gelling ability of this important plant polysaccharide for the food industry. Plant apoplastic pectinacetylesterases (PAEs) play a key role in regulating the degree of pectin acetylation and modifying their expression thus represents one way to engineer plant polysaccharides for food applications. Identifying the major active enzymes within the PAE gene family will aid in our understanding of this biological phenomena as well as provide the tools for direct trait manipulation. Using comparative genomics we propose that there is a minimal set of 4 distinct PAEs in plants. Possible functional diversification of the PAE family in the grasses is also explored with the identification of 3 groups of PAE genes specific to grasses.
Keywords: acetylation; cell walls; comparative genomics; pectin; pectinacetylesterases; phylogeny.
Figures

Similar articles
-
Plant pectin acetylesterase structure and function: new insights from bioinformatic analysis.BMC Genomics. 2017 Jun 8;18(1):456. doi: 10.1186/s12864-017-3833-0. BMC Genomics. 2017. PMID: 28595570 Free PMC article.
-
Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants.Planta. 2014 Nov;240(5):1123-38. doi: 10.1007/s00425-014-2139-6. Epub 2014 Aug 13. Planta. 2014. PMID: 25115560 Free PMC article.
-
Pectin-modifying enzymes and pectin-derived materials: applications and impacts.Appl Microbiol Biotechnol. 2014 Jan;98(2):519-32. doi: 10.1007/s00253-013-5388-6. Epub 2013 Nov 24. Appl Microbiol Biotechnol. 2014. PMID: 24270894 Review.
-
Microbial carbohydrate esterases deacetylating plant polysaccharides.Biotechnol Adv. 2012 Nov-Dec;30(6):1575-88. doi: 10.1016/j.biotechadv.2012.04.010. Epub 2012 May 9. Biotechnol Adv. 2012. PMID: 22580218 Review.
-
A Bacillus licheniformis pectin acetylesterase is specific for homogalacturonans acetylated at O-3.Carbohydr Polym. 2014 Jul 17;107:85-93. doi: 10.1016/j.carbpol.2014.02.006. Epub 2014 Feb 13. Carbohydr Polym. 2014. PMID: 24702921
Cited by
-
The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs).Int J Mol Sci. 2018 Sep 21;19(10):2878. doi: 10.3390/ijms19102878. Int J Mol Sci. 2018. PMID: 30248977 Free PMC article. Review.
-
Bioinformatics analysis of PAE family in Populus trichocarpa and responsiveness to carbon and nitrogen treatment.3 Biotech. 2021 Aug;11(8):370. doi: 10.1007/s13205-021-02918-1. Epub 2021 Jul 13. 3 Biotech. 2021. PMID: 34295610 Free PMC article.
-
Inhibition of peroxidases and oxidoreductases is crucial for avoiding false-positive reactions in the localization of reactive oxygen species in intact barley root tips.Planta. 2022 Feb 16;255(3):69. doi: 10.1007/s00425-022-03850-1. Planta. 2022. PMID: 35174422
-
Plant Cell Wall Polysaccharide O-Acetyltransferases.Plants (Basel). 2024 Aug 19;13(16):2304. doi: 10.3390/plants13162304. Plants (Basel). 2024. PMID: 39204739 Free PMC article. Review.
-
Systematic Analysis and Functional Validation of Citrus Pectin Acetylesterases (CsPAEs) Reveals that CsPAE2 Negatively Regulates Citrus Bacterial Canker Development.Int J Mol Sci. 2020 Dec 11;21(24):9429. doi: 10.3390/ijms21249429. Int J Mol Sci. 2020. PMID: 33322321 Free PMC article.
References
-
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant Cell Walls: From Chemistry to Biology. New York: Garland Science, 2011.
-
- Atmodjo MA, Hao ZY, Mohnen D. Evolving views of pectin biosynthesis. Annu Rev Plant Biol 2013; 64:747-79; PMID:23451775; http://dx.doi.org/10.1146/annurev-arplant-042811-105534 - DOI - PubMed
-
- Harholt J, Suttangkakul A, Scheller HV. Biosynthesis of pectin. Plant Physiol 2010; 153:384-95; PMID:20427466; http://dx.doi.org/10.1104/pp.110.156588 - DOI - PMC - PubMed
-
- de Souza A, Hull PA, Gille S, Pauly M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 2014; 240:1123-38; PMID:25115560; http://dx.doi.org/10.1007/s00425-014-2139-6 - DOI - PMC - PubMed
-
- Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. The Plant Cell 2012; 24:50-65; PMID:22247250; http://dx.doi.org/10.1105/tpc.111.092411 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
