A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance
- PMID: 26238195
- PMCID: PMC4523026
- DOI: 10.1186/s12864-015-1778-8
A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance
Abstract
Background: Chickens are susceptible to infection with a limited number of Influenza A viruses and are a potential source of a human influenza pandemic. In particular, H5 and H7 haemagglutinin subtypes can evolve from low to highly pathogenic strains in gallinaceous poultry. Ducks on the other hand are a natural reservoir for these viruses and are able to withstand most avian influenza strains.
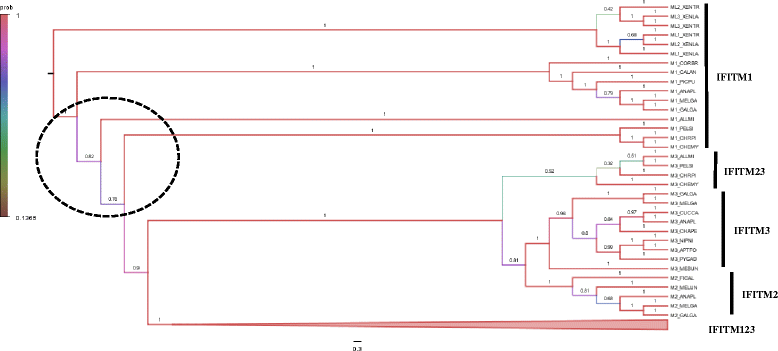
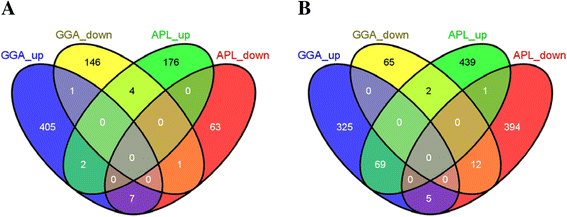
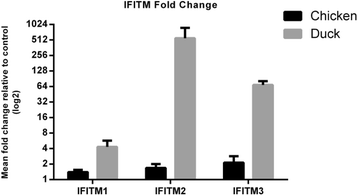
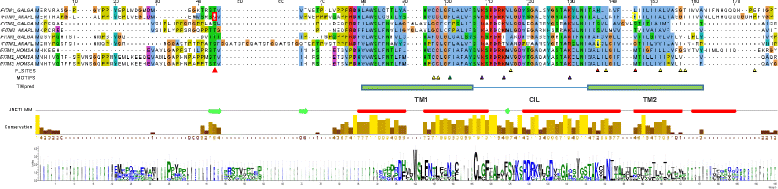
Results: Transcriptomic sequencing of lung and ileum tissue samples from birds infected with high (H5N1) and low (H5N2) pathogenic influenza viruses has allowed us to compare the early host response to these infections in both these species. Chickens (but not ducks) lack the intracellular receptor for viral ssRNA, RIG-I and the gene for an important RIG-I binding protein, RNF135. These differences in gene content partly explain the differences in host responses to low pathogenic and highly pathogenic avian influenza virus in chicken and ducks. We reveal very different patterns of expression of members of the interferon-induced transmembrane protein (IFITM) gene family in ducks and chickens. In ducks, IFITM1, 2 and 3 are strongly up regulated in response to highly pathogenic avian influenza, where little response is seen in chickens. Clustering of gene expression profiles suggests IFITM1 and 2 have an anti-viral response and IFITM3 may restrict avian influenza virus through cell membrane fusion. We also show, through molecular phylogenetic analyses, that avian IFITM1 and IFITM3 genes have been subject to both episodic and pervasive positive selection at specific codons. In particular, avian IFITM1 showed evidence of positive selection in the duck lineage at sites known to restrict influenza virus infection.
Conclusions: Taken together these results support a model where the IFITM123 protein family and RIG-I all play a crucial role in the tolerance of ducks to highly pathogenic and low pathogenic strains of avian influenza viruses when compared to the chicken.
Figures



References
-
- Kajihara M, Sakoda Y, Soda K, Minari K, Okamatsu M, Takada A, et al. The PB2, PA, HA, NP, and NS genes of a highly pathogenic avian influenza virus A/whooper swan/Mongolia/3/2005 (H5N1) are responsible for pathogenicity in ducks. Virol J. 2013;10:45. doi: 10.1186/1743-422X-10-45. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

