Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2
- PMID: 26238318
- PMCID: PMC4532830
- DOI: 10.1038/ncomms8871
Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2
Abstract
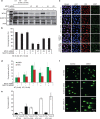
Deregulated redox metabolism in cancer leads to oxidative damage to cellular components including deoxyribonucleoside triphosphates (dNTPs). Targeting dNTP pool sanitizing enzymes, such as MTH1, is a highly promising anticancer strategy. The MTH2 protein, known as NUDT15, is described as the second human homologue of bacterial MutT with 8-oxo-dGTPase activity. We present the first NUDT15 crystal structure and demonstrate that NUDT15 prefers other nucleotide substrates over 8-oxo-dGTP. Key structural features are identified that explain different substrate preferences for NUDT15 and MTH1. We find that depletion of NUDT15 has no effect on incorporation of 8-oxo-dGTP into DNA and does not impact cancer cell survival in cell lines tested. NUDT17 and NUDT18 were also profiled and found to have far less activity than MTH1 against oxidized nucleotides. We show that NUDT15 is not a biologically relevant 8-oxo-dGTPase, and that MTH1 is the most prominent sanitizer of the cellular dNTP pool known to date.
Figures

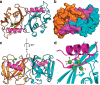


References
-
- Cheung-Ong K., Giaever G. & Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem. Biol. 20, 648–659 (2013). - PubMed
-
- Bouwman P. & Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 12, 587–598 (2012). - PubMed
-
- Rai P. et al. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene 30, 1489–1496 (2011). - PubMed
-
- Zhang Y. et al. Redox control of the survival of healthy and diseased cells. Antioxid. Redox Signal 15, 2867–2908 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

